-
食品加工、制药工业、皮革工业、垃圾渗滤液等废水不仅氨氮 (NH4+-N) 浓度高,还含有大量盐分[1-3]。生物法虽然作为一种经济有效的废水处理方法,但是高盐分可抑制微生物酶活,甚至引起细胞质壁分离导致微生物死亡[4-5],不利于微生物对NH4+-N的降解。因此,在对高盐废水的脱氮处理中,通常使用膜分离[6]、蒸发结晶[7]、芬顿[8]和电渗析技术[9]等物化法进行脱盐预处理,这不但使运行成本高、工艺流程长,还会造成二次污染。虽然经物化脱盐处理后的高盐含氮废水可进一步通过生物法脱氮,但去除效果并不稳定[10-11],即使在低盐度环境(<2%),传统硝化、反硝化菌和常规异养菌等常见脱氮微生物的生长代谢及活性也会受到明显抑制[12-13]。具备异养硝化-好氧反硝化(heterotrophic nitrification-aerobic denitrification, HN-AD)功能的好氧脱氮菌在好氧条件下因具备同时硝化反硝化、且无中间产物亚硝态氮 (NO2−-N) 和硝态氮 (NO3−-N) 积累的特质得到广泛关注[14]。不仅如此,部分HN-AD菌属还兼具耐盐性,已被广泛应用于高盐废水处理过程,如不动杆菌属 (Acinetobacter) 、盐单胞菌属 (Halomonas) 、副球菌属 (Paracoccus) [15]等。然而,在工程应用中,许多工业废水的盐度波动大,这使得包括HN-AD菌在内的众多微生物难以快速适应不同的盐度环境,菌群无法实现高效富集,进而影响废水处理效果[16-17]。为了增强微生物对高盐胁迫的抵抗能力,已有研究[18]通过增加盐浓度驯化功能菌属耐盐性能的方式以期获得高去污效率、高富集丰度的耐盐型脱氮功能微生物。然而,在实际应用中却发现,即使经过长期驯化,功能微生物对高盐胁迫的抵抗能力仍较弱[19-20],原因在于功能微生物易流失、难截留导致富集丰度较低,最终影响废水处理效果的稳定性[19, 21-22]。因此,为解决这一问题,亟待研发一种即可提高微生物对高盐胁迫的抵抗能力,又可提高功能菌属生长富集能力的新技术。
微生物在逆境中可通过积累或从环境中吸收相容性溶质 (compatible solutes, CS) 来抵抗外部条件带来的胁迫作用,在不需要细胞内酶的任何特殊适应下即可保护细胞免受不可逆转的损害[23-24]。所以CS在细胞中发挥了重要作用,它不仅不会阻碍正常的细胞代谢,还可以平衡细胞质内的渗透压以增强微生物的活性[23, 25]。在VYRIDES等[26]的研究中,盐度为4%时,假单胞菌株 (Pseudomonas sp strain ADP) 产生的相容性溶质主要为海藻糖;ZHANG等[27]在使用颗粒污泥处理含盐养殖废水的过程中,添加1 000 μmol·L−1的海藻糖,可使产甲烷活性和磷酸酶活性分别提高22%和27%。而海藻糖作为一种价格低、易采购的典型相容性溶质,在高盐废水处理中已被成功应用[28-29]。其作为渗透调节剂易被微生物吸收利用,通过调节细胞渗透压帮助微生物快速适应高盐环境,提升去污能力[26]。目前,已有海藻糖用于高盐废水的厌氧处理系统的相关报道[29-30]。BAI等[30]研究了在序批式反应器中添加海藻糖考察了厌氧氨氧化细菌处理富氮含盐废水的长期脱氮性能,在添加浓度为300 μmol·L−1时,就可使NH4+-N、NO2−-N的去除率分别提高为32.4%和42.2%;杨振琳等[28]通过添加海藻糖强化厌氧化耦合反硝化工艺,在盐度为3.2%的模拟废水中加入250 μmol·L−1的海藻糖,NH4+-N和NO2−-N的去除速率分别提高了81.25%和75%。可见海藻糖对提高长期脱氮效果有显著性作用,但其在高盐废水好氧生物处理中的研究却鲜有报道。盐度显著影响微生物的物理和生化特性,长期作用甚至能够改变整个环境的微生物群落结构[31]。
本研究利用膜曝气生物膜反应器 (membrane aerobic biofilm reactor, MABR) 富集好氧脱氮菌,通过调节外加海藻糖的浓度,采用高通量测序等分析方法,从微生物脱氮性能、群落结构及脱氮功能基因丰度等层面,研究海藻糖浓度梯度对高盐废水中微生物的物种丰度、菌群多样性及脱氮功能基因的影响,旨在为推动好氧脱氮菌在高盐废水好氧生物脱氮处理的实际应用提供理论基础及新技术思路。
-
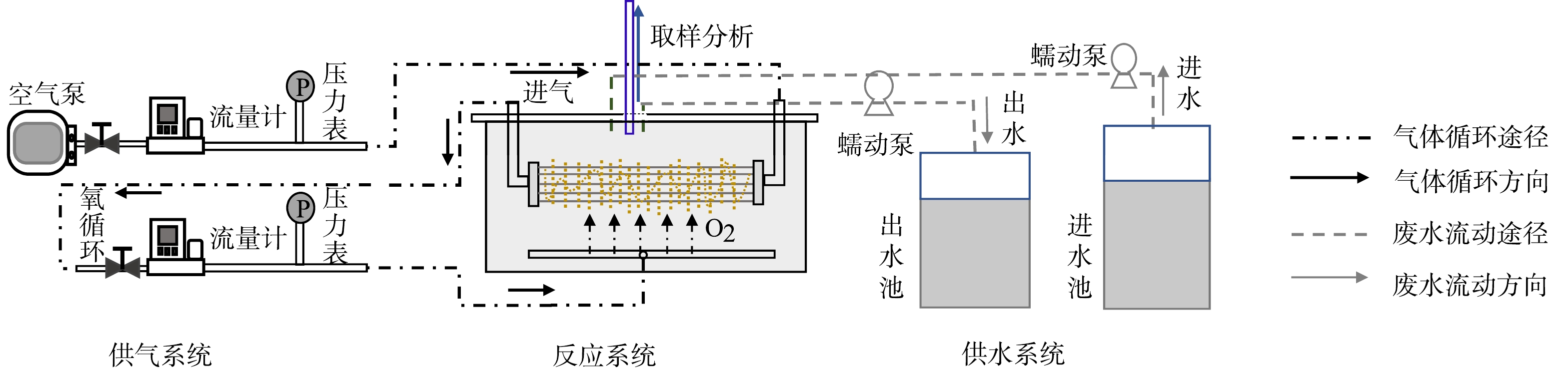
在实验过程中,建立了5个MABR,工作体积为0.85 L,反应装置见图1。反应器的膜组件由128根中空纤维膜组成,用于空气运输和生物膜附着,中空纤维是用复合聚合物材料制造,只允许气体分子渗透。纤维的外径为300 μm,内径为200 μm,膜孔径为0.1 μm,有效比表面积为1.76 cm2·cm−3。膜组件一端通过流量计和压力表连接到空气泵,另一端连接到底端曝气管形成氧循环,曝气压力设定为10 kPa,进出水均采用蠕动泵完成,水力停留时间为48 h[32]。反应器顶部留有取样口,用于采样和溶解氧 (DO) 监测等操作。
-
合成的高盐高氮废水由无水CH3COONa (碳源) 、(NH4)2SO4 (氮源) 、NaCl和微量元素组成。进水COD为5 000 mg·L−1,NH4+-N为500 mg·L−1,盐度为2.5%,微量元素为50 mL·L−1。微量元素配方: MgSO4·7H2O 2.0 g·L−1,MnSO4·H2O 0.1 g·L−1,CaCl2 1.5 g·L−1,FeSO4·7H2O 0.1 g·L−1,K2HPO4 5.0 g·L−1。
该研究的接种菌液中主要为产碱杆菌(Alcaligenes)、不动杆菌(Acinetobacter sp.TAC-1)和假单胞菌 (Pseudomonas) 等具备HN-AD功能的好氧脱氮混合菌群。这些细菌主要分离于猪场沼液、垃圾渗滤液和化工废水,可耐受高浓度氨氮。为确保适当的菌液浓度和脱氮活性,首先进行菌液驯化,再分别接种于5个反应器中进行挂膜。挂膜完成后,分别加入不同浓度海藻糖 (0、40、120、360和1 080 μmol·L−1) 进行实验,对应反应器分别为C0、C40、C120、C360和C1 080。
-
反应器启动运行阶段的溶氧、盐度及pH值分别采用便携式溶氧仪、海水比重计及pH计进行直接测量。菌液浓度 (OD600 nm) 采用紫外分光光度法测定,进、出水NH4+-N、NO3−-N、NO2−-N、TN和COD的浓度均参照文献中的方法[33]进行检测。
利用扫描电镜 (scanning electron microscope, SEM) 技术表征反应器挂膜阶段的生物膜表观形态特征。将采集的生物膜样品经过固定、PBS清洗、乙醇梯度脱水 (30%、50%、70%、85%和95%质量分数的乙醇洗脱1次,100%乙醇洗脱2次(每次脱水时间为15~20 min)) 、冷冻干燥等步骤完成样品预处理,将制备完成的样品送至武汉铄思百检测技术有限公司并进行SEM观察。
-
在各反应器稳定运行阶段,采集微生物样品(每个反应器重复采集3次样品,每个样品约0.5 g),将样品短期保存于−80 ℃冰箱中。采用通用检测试剂盒 (MOBIO实验室,美国) 提取样品的总基因组DNA,采用引物338F和806R扩增V3~V4区域的l6S rRNA基因,测序引物序列为338F(5'-ACTCCTACGGGAGGCAGCA-3')和806R(5'-GGACTACHVGGGTWTCTAAT-3')。采用Trimmomatic和Flash软件对测序序列进行拼接,通过Usearch 软件对具有97%相似性的序列进行OTU聚类,经Usearch_global 获得OTU序列丰度统计表,进行后续微生物群落多样性分析;基于京都基因和基因组百科全书(http://www.genome.jp·KEGG)数据库及PICRUSt1软件,对样品中微生物脱氮功能基因进行预测分析。
-
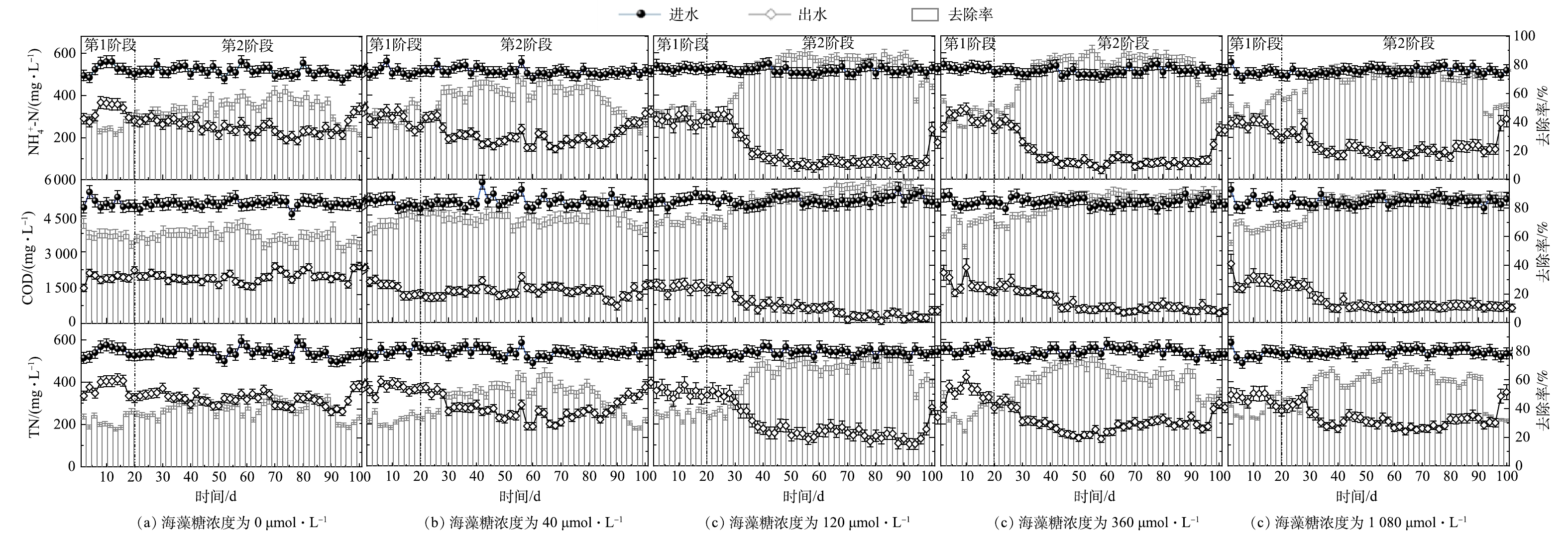
SEM表征结果显示,反应器启动运行20 d,膜丝上明显附着了大量的以杆菌和球菌为主的微生物 (图2) ,说明反应器于20 d内成功完成挂膜启动。该研究反应器菌液挂膜时间较文献报道相关结果[32-34]略有增加,这可能是因为盐度抑制了微生物的生长繁殖,从而导致挂膜周期延长。反应器启动运行阶段的污染物去除情况如图3(a)~图3(e)中第1阶段所示。5个平行反应器的NH4+-N、TN及COD去除率分别为32%~45%、32%~40%和60%~71%,无明显有害中间产物NO2−-N和NO3−-N的积累。启动阶段反应器脱氮性能较低,究其原因主要在于高盐环境对反应器富集的脱氮微生物的生长及活性存在影响。高盐度会使微生物细胞处于一个高渗环境,影响细胞正常生长代谢,进而降低污染物的去除率[35]。
在4个平行反应器中添加不同浓度海藻糖 (40、120、360和1 080 μmol·L−1) ,设定为实验组 (C40、C120、C360和C1 080) ,对照组 (C0) 不添加海藻糖,连续运行80 d结束。添加海藻糖后的反应器运行性能如图3(a)~图3(e)中第2阶段所示。相较于对照组,4个实验组中NH4+-N去除率分别提高了10.70%、32.72%、27.36%、19.45%,TN去除率分别提高了8.32%、28.36%、22.53%、17.63%,COD去除率分别提高了12.09%、31.14%、25.27%、25.06%。这说明添加海藻糖对高盐给微生物带来的高渗胁迫具有缓解作用,提高了系统脱氮性能。C120组的NH4+-N、TN及COD去除率最高,C360组的TN去除率及C1 080组的NH4+-N、TN去除率相较C120组降低,3组中COD去除率无明显变化。可以看出,外源海藻糖浓度与污染物去除率无线性相关性,最适海藻糖浓度为120 μmol·L−1。这主要是因为微生物对海藻糖的最大渗透调节剂有一定的吸收范围[28],当体系内海藻糖浓度超过微生物作为渗透调节剂的吸收浓度范围时,过剩的海藻糖可充当常规异养反硝化菌 (heterotrophic denitrified bacteria, HDB) 的有机碳源[28]。因此,在本研究中,高浓度海藻糖 (360、1 080 μmol·L−1) 条件可能对体系内HDB生长更有利,进而影响好氧脱氮菌群的富集与活性,降低了反应器脱氮性能。
-
1) 微生物α多样性分析。通过微生物群落分析研究不同海藻糖浓度对反应器中物种丰富度及多样性的影响,结果如表1所示。通过MiSeq Illumina测序分析获得269~303个OTU,覆盖指数均大于99%,说明测序深度包含测序样品中的全部细菌数。Ace指数和Chao指数均为物种丰富度的指标,实验组较对照组有所增加,表明添加海藻糖提升了高盐环境下系统内的物种丰富度。Shannon指数反映微生物多样性特征,其数值同样在实验组中有所增长,说明添加海藻糖丰富了高盐环境下系统内的菌群多样性。C120组的Chao指数、Ace指数以及Shannon指数均最高,说明120 μmol·L−1的海藻糖浓度对增强高盐胁迫下微生物群落的抵抗力稳定性最有利。
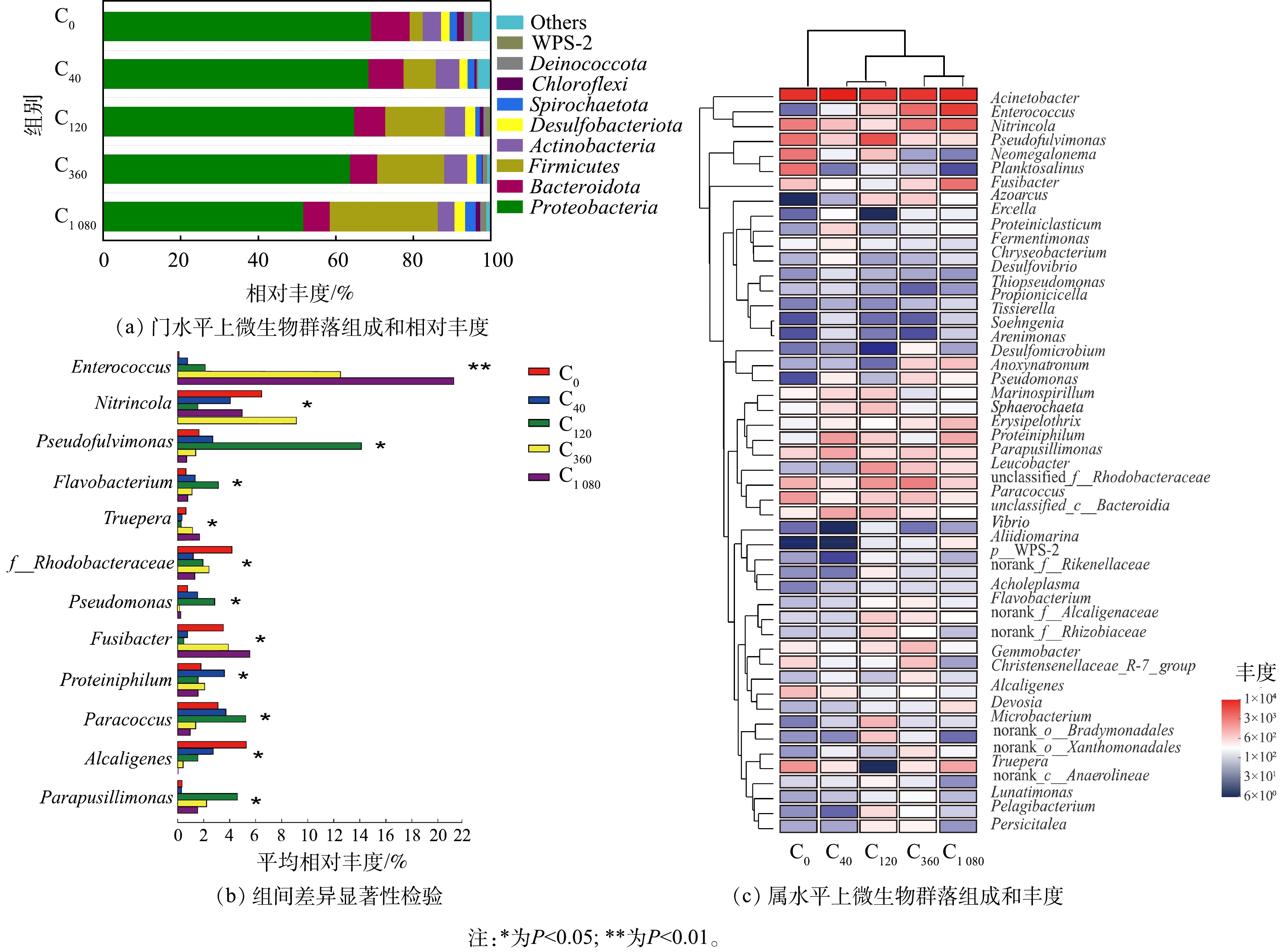
2) 微生物群落组成分析。不同海藻糖浓度下微生物群落组成相似但相对丰度略有差异(图4)。如图4(a)所示,门水平相对丰度排名前3的优势菌门分别为变形菌(Proteobacteria)、拟杆菌(Bacteroidota)和厚壁菌(Firmicutes)。三者的总相对丰度在对照组C0中为82.60%,在各实验组 (C40、C120、C360和C1 080) 中分别提高了3.29%、5.64%、5.49%和3.84%,说明在高盐条件下添加海藻糖对系统中门水平的群落结构具有一定影响。有报道[36-37]指出,Proteobacteria和Bacteroidota在高盐、高氨氮废水处理中常占据主导地位,其包含着多样性最丰富的硝化和反硝化细菌 (如HN-AD菌) ;也有报道[38]指出,可在极端环境进行异养硝化的脱氮微生物 (如肠球菌属Enterococcus) 归属于Firmicutes。该3类菌门中微生物总相对丰度的增加说明添加海藻糖有利于高盐胁迫下脱氮菌群的生长富集。
由属水平相对丰度排名前50的微生物进行层级聚类Heatmap分析结果 (图4(c)) 可知,在C0组中,不动杆菌属 (Acinetobacter) 相对丰度为34.58%,是系统内的优势菌属,Acinetobacter在高氨氮浓度下是硝化和反硝化过程中生长的主要菌群,表现出高效的HN-AD能力[39],且具有耐受高盐、高氨氮和脱氮速率快的优点[40]。而添加海藻糖的4个实验组中,高丰度优势菌属仍为Acinetobacter且相对丰度保持稳定(分别为35.69%、34.95%、34.93%和31.04%)。这说明在高盐环境下,海藻糖对Acinetobacter生长富集的影响较弱,但Acinetobacter保证了高盐系统内的高脱氮效率。在C120组中,假黄褐藻属 (Pseudofulvimonas) 相对丰度明显高于对照组和其他实验组,并且其相丰度变化存在显著性差异 (P <0.05) (图4(b)) 。Pseudofulvimonas相对丰度在低浓度海藻糖时 (40、120 μmol·L−1),随海藻糖浓度的增加而增长,但在高浓度海藻糖时 (360、1 080 μmol·L−1) ,却随海藻糖浓度的增加而减少。Pseudofulvimonas为耐盐型HN-AD菌,是高盐废水处理过程中常见的脱氮功能微生物[41-42];同时,黄杆菌属 (Flavobacterium) [43]、红杆菌属 (Rhodobacteraceae) [44]、假单胞菌属 (Pseudomonas) [16]、副球菌属 (Paracoccus) [45]和产碱菌科 (Parapusillimonas) [46]等耐盐型HN-AD菌在海藻糖浓度作用下的相对丰度变化规律与Pseudofulvimonas一致并同样存在显著性差异 (P<0.05) (见图4(b)) 。Pseudofulvimonas通常与各种氮物质降解过程有关[47];Flavobacterium与生物絮体聚集和颗粒稳定性有关,其丰度增加有利于功能微生物的富集进而保证污染物稳定有效的去除[42, 48];Rhodobacteraceae是海水形成初始生物膜的主要定殖者[44],其具有较强的耐盐性能,能将NH4+-N转化为NO3−-N和NO2−-N,且可在曝气的条件下实现对NO3−-N的高效去除[49-50];Pseudomonas、Paracoccus具有亚硝酸盐还原和有机物降解等多种功能,在有机物去除方面起着关键作用[51-52]。当海藻糖浓度为120 μmol·L−1时,这些HN-AD菌属相对丰度均达到最高。说明低浓度海藻糖促进高盐废水中耐盐型HN-AD菌生长富集,使系统内主要发生异养硝化和好氧反硝化过程,可强化微生物对氮、碳污染物的去除性能。
在C360和C1 080组中,Enterococcus相对丰度由C0中的0.12%分别增至13.88%和21.22% (P <0.05) (图4(b)) 。Enterococcus是一种兼性脱氮菌[53],具有较强的脱氮耐盐性能及高效的产絮凝效果,在生长代谢过程中产生多糖、蛋白质、脂和核酸等成分,可促进水体中菌体细胞凝聚进而减缓功能微生物的流失[54-55]。Enterococcus相对丰度在C360和C1 080组得到明显提升,说明高浓度海藻糖的作用倾向于通过增强高盐条件下微生物凝聚能力,提高功能菌群富集。此外,硝化细菌属 (Nitrincola) 、特吕珀菌属 (Truepera) 和梭菌属 (Fusibacter) 3种HDB的相对丰度在高浓度海藻糖时 (360、1 080 μmol·L−1) 也呈现增长趋势。Nitrincola是一类耐盐碱的厌氧反硝化细菌[15],能够在极端环境中对地球化学元素循环起促进作用[56];Truepera[32]与厌氧反硝化作用密切相关[57],且能够在碱性、中等盐度和高温等多种极端条件下生长,有利于NO向N2O的转换[58-59];Fusibacter是一类耐盐、厌氧的棒杆细菌,可将硫酸盐和元素硫还原成硫化物,还能够广泛利用碳水化合物 (如乙酸、葡萄糖等) 生成CO2和H2等产物[60],且具有脱氮除磷的功能[61]。上述3种菌属均在高浓度氨氮环境的反硝化过程中发挥重要作用,其相对丰度在高海藻糖浓度作用下提高,也证实高浓度海藻糖促进高盐废水中HDB生长富集,使系统内主要发生常规异养反硝化过程。这主要是因为高浓度海藻糖促进Enterococcus相对丰度大幅增加,虽有利于微生物凝聚,但同时也产生大量菌胶团聚集堵塞膜孔,致使生物膜层出现缺氧环境,不利于好氧脱氮微生物的生长,继而引起HDB增殖。虽然HDB相对丰度在高海藻糖浓度下的增长促使C360和C1 080组的NH4+-N和TN去除率相比C0组有所提高,但其相比C40和C120组却呈下降趋势,推测耐盐型HN-AD菌的相对丰度变化对系统脱氮性能的影响相比HDB更为显著和重要,而HDB的相对丰度变化主要影响了系统的脱氮菌群结构。
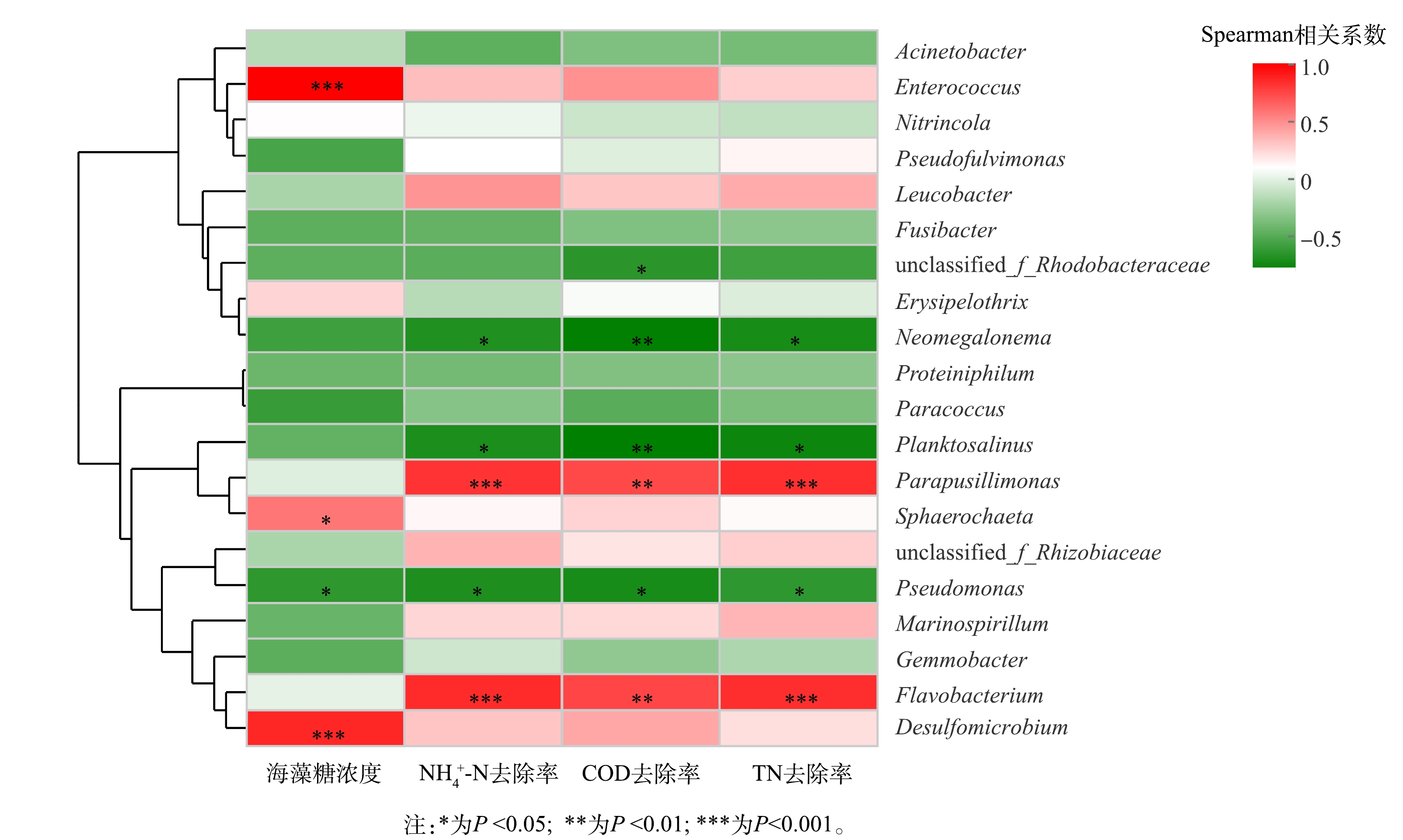
3) 环境因子相关性分析。通过相关性Heatmap图进一步分析环境因子对系统内属水平群落组成的影响,结果如图5所示。与NH4+-N、COD及TN去除率和海藻糖浓度4个因子均显著关联的只有Pseudomonas的相对丰度,且呈负相关;NH4+-N、COD及TN去除率与Flavobacterium、Parapusillimonas 2种耐盐型HN-AD菌属相对丰度呈正相关,与浮游盐属 (Planktosalinus) 、新巨藻属(Neomegalonema)相对丰度呈负相关。Planktosalinus归属黄杆菌科 (Flavobacteriaceae) ,而Flavobacteriaceae是废水系统发生污泥膨胀的指示微生物[62-63];Neomegalonema同样属于污泥膨胀菌群,其丰度变化用于表征氮磷失衡下废水中的微生物群落健康状况[64-65]。海藻糖浓度与Enterococcus、球毛菌属 (Sphaerochaeta) 和脱硫微杆菌属 (Desulfomicrobium) 相对丰度呈正相关。可以看出,不同菌属相对丰度受到海藻糖浓度、污染物去除率影响的情况存在差异,可证明耐盐型HN-AD菌确为系统内主要脱氮贡献者,显著影响反应器的脱氮性能;以Enterococcus为代表的传统脱氮菌和其他功能微生物的相对丰度变化规律则主要反映了海藻糖浓度梯度对高盐废水中脱氮菌群组成的差异影响。
-
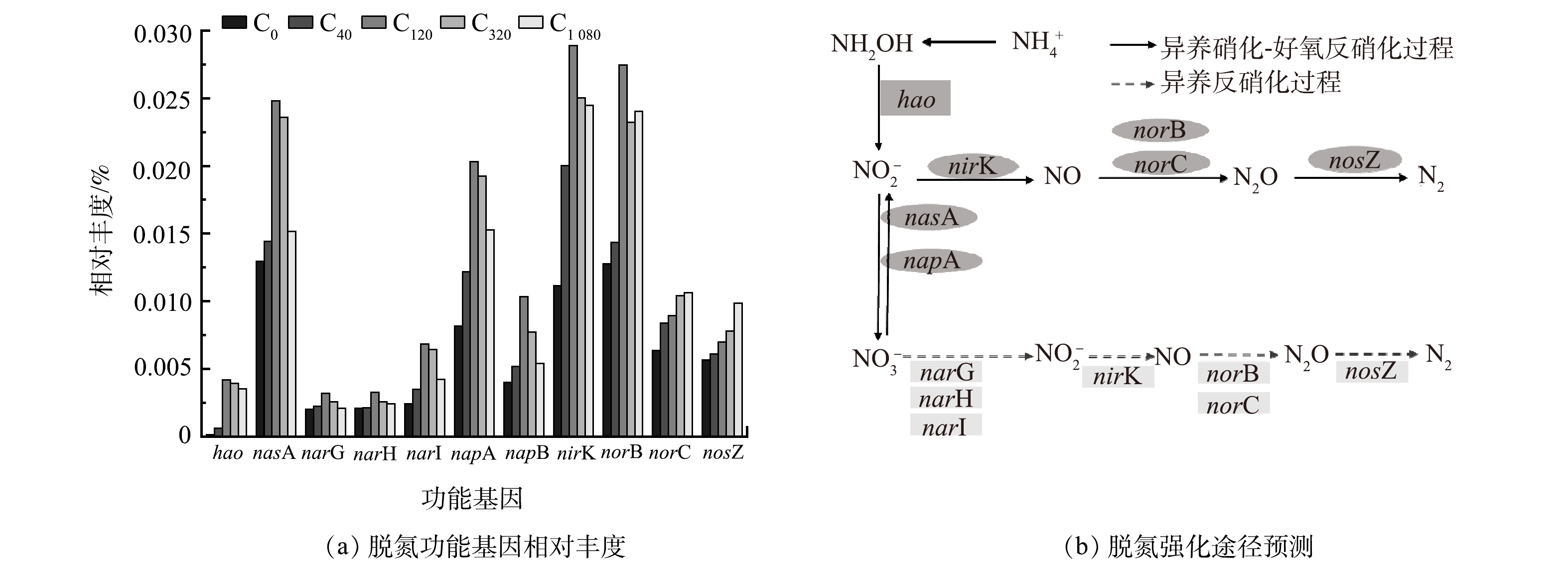
采用PICRUSt1对功能基因丰度进行分析,其相对丰度变化结果见图6(a)。该研究获得与硝化过程相关的羟胺氧化酶基因hao,是NH2OH氧化生成NO2−-N过程的关键酶基因[66],其在实验组 (C40、C120、C360和C1 080) 中相对丰度相较对照组 (C0) 分别提高了4.96倍、34.70倍、32.45倍和29.06倍,说明添加海藻糖明显加快系统内微生物的硝化进程。同时,还获得8个与反硝化过程相关的功能基因,包括硝酸盐还原酶(nasA、nasB、narG、narH、narI、napA、napB)、亚硝酸盐还原酶(nirK)、一氧化氮氧化酶(norB、norC)和一氧化二氮还原酶(nosZ)。4个实验组中反硝化功能基因总相对丰度较对照组分别提高了1.31倍、2.86倍、1.81倍和1.75倍,说明添加海藻糖同样促进了系统内微生物的反硝化进程。
硝酸还原酶催化细菌体内NO3−-N还原生成NO2−-N,是实现反硝化过程的关键酶之一。nasA是一种同化型硝酸盐还原酶基因,存在于细菌的细胞质中[67],也常见于HN-AD菌[68]。其在实验组中的相对丰度值相较对照组均有提高,低浓度海藻糖时 (40、120 μmol·L−1) ,nasA基因相对丰度随海藻糖浓度的增加而增长,于海藻糖浓度为120 μmol·L−1时达到最高值;但在高浓度海藻糖时 (360、1 080 μmol·L−1) ,其随海藻糖浓度增加出现降低。同样,异化型硝酸盐还原酶NAP的基因napA和napB相对丰度变化规律与nasA一致。NAP表达在好氧条件下占主导地位,常存于HN-AD菌中[69]。另一种异化型硝酸还原酶NAR的基因narG、narH和narI相对丰度却在海藻糖浓度为360 μmol·L−1和1 080 μmol·L−1时增加,而NAR表达在缺氧条件下占主导地位,常见于HDB[70]。以上结果说明,在高盐废水添加海藻糖可诱导增强以HN-AD菌和HDB为主的脱氮菌群的反硝化活性。低浓度海藻糖倾向于强化系统的异养硝化-好氧反硝化途径,而高浓度海藻糖则偏向于促进系统的异养反硝化途径 (图6(b)) 。
亚硝酸盐还原酶NIR是反硝化过程的另一类关键酶,驱动NO2−-N还原生成气体产物NO,存在于细胞膜外生物细胞质中,其基因有nirS和nirK 2种类型[71]。海藻糖的加入提高了nirK的相对丰度,且在海藻糖浓度为120 μmol·L−1时达到最高值,推进了系统的亚硝酸盐还原进程。NO还原酶基因 (norB和norC) 和nosZ分别是催化NO还原生成N2O、N2O进一步还原生成最终产物N2的关键酶基因。在污水处理中,反硝化细菌体内nosZ活性的降低或不具有nosZ是导致反硝化过程中温室气体N2O大量积累的主要因素[72]。该研究nosZ的相对丰度随着海藻糖浓度的增加而升高,说明添加海藻糖对控制好氧脱氮过程的N2O产生与积累具有一定作用。以上基因预测结果也进一步证明,海藻糖浓度为120 μmol·L−1时,反应器的NH4+-N、TN去除率最高 (图2) 。这主要是因为此海藻糖浓度条件激发HN-AD菌的硝化和反硝化进程达到最快,极大程度加速了NH4+-N氧化及NO3−-N和NO2−-N的还原,最终使反应器脱氮性能达到最优。
-
1) 添加海藻糖的实验组 (40、120、360和1 080 μmol·L−1) 中NH4+-N、TN和COD去除率相较对照组 (0 μmol·L−1) 显著提高,最适海藻糖浓度为120 μmol·L−1。外加海藻糖可优化高盐条件下的反应器脱氮性能。
2) 海藻糖浓度对脱氮菌群的相对丰度具有显著影响。低浓度海藻糖 (40、120 μmol·L−1) 有利于耐盐型HN-AD菌相对丰度的提高,而高浓度海藻糖 (360、1 080 μmol·L−1) 更倾向于促进HDB的富集;耐盐型HN-AD菌是系统的主要脱氮贡献者,其相对丰度变化与污染物去除率之间存在显著相关性,传统脱氮菌相对丰度变化则与高盐废水中脱氮菌群组成差异有关。耐盐型HN-AD菌对调控高盐条件下的反应器脱氮性能起着重要影响。
3) 海藻糖可有效提高以HN-AD菌和HDB为主的脱氮菌群的硝化和反硝化活性。在低浓度海藻糖 (40、120 μmol·L−1) 作用下,与HN-AD菌关联的硝化、反硝化基因(hao、nasA、napA和napB)丰度随海藻糖浓度的增加而增长,系统的异养硝化-好氧反硝化途径得到强化;在高浓度海藻糖 (360、1 080 μmol·L−1) 作用下,与HDB关联的反硝化基因 (narG、narH和narI) 相对丰度明显增加,系统的异养反硝化途径得到增强。
外源性海藻糖强化好氧脱氮菌处理高盐废水的菌群特性
Characterization of trehalose enhanced aerobic denitrifying bacteria for high-salt wastewater treatment
-
摘要: 为提升高盐胁迫下好氧脱氮功能菌群的富集丰度及脱氮性能,解决高盐废水生物脱氮效率低的问题,助推好氧脱氮功能菌在高盐废水处理的工程应用,将海藻糖添加到已富集好氧脱氮功能菌的膜曝气生物膜反应器 (membrane aerobic biofilm reactor, MABR) 中,构建高盐废水的海藻糖生物强化处理系统,从反应器脱氮性能、菌群多样性以及脱氮功能基因丰度等方面探究高盐废水中海藻糖对好氧脱氮菌的强化机制。结果表明:实验组 (C40、C120、C360和C1 080) 中NH4+-N、TN和COD去除率相较对照组 (C0) 分别提高了10.70%、32.72%、27.36%、19.45%,8.32%、28.36%、22.53%、17.63%和12.09%、31.14%、25.27%、25.06%;外加海藻糖提高了菌群在高盐废水中的脱氮效率,浓度为120 μmol·L-1时,NH4+-N去除率最高可提升32.72%。高通量测序分析显示:在高盐环境胁迫下,海藻糖浓度对群落结构及丰度存在显著影响,C40和C120提高了异养硝化-好氧反硝化 (heterotrophic nitrification-aerobic denitrification, HN-AD) 菌 (Pseudofulvimonas、Rhodobacteraceae、 Paracoccus、Parapusillimonas和Flavobacterium) 的相对丰度,而C360和C1 080有利于异养反硝化菌 (heterotrophic denitrified bacteria, HDB) (Enterococcu、Nitrincola、Truepera、Fusibacter) 的富集;海藻糖浓度显著影响高盐废水中脱氮菌群的组成与丰度。PICRUSt1结果显示:添加海藻糖有效提高以HN-AD菌和HDB为主的脱氮菌群的反硝化活性,在C40和C120中,与HN-AD菌关联的硝化基因 (hao) 和反硝化基因 (nasA、napA和napB) 相对丰度提高,此时HN-AD途径得到增强;而与HDB关联的反硝化基因 (narG、narH和narI) 相对丰度在C360和C1 080增加,进一步说明加入高浓度海藻糖更有利于加强异养反硝化途径;海藻糖浓度为120 μmol·L-1时,脱氮基因相对丰度最高,最大程度地加快了好氧脱氮菌群的硝化和反硝化进程。本研究结果可为好氧脱氮菌在高盐废水的好氧处理技术运用中提供参考。
-
关键词:
- 海藻糖 /
- 异养硝化-好氧反硝化 /
- 高盐废水 /
- 微生物多样性 /
- 功能基因
Abstract: In order to enhance the enrichment abundance and denitrification performance of aerobic denitrifying functional bacteria under high-salt stress, which can solve the problem of low efficiencies of biological denitrification of high-salt wastewater and promote the engineering application of aerobic denitrifying functional bacteria in high-salt wastewater treatment, the trehalose was added to the membrane aerobic biofilm reactor (MABR) with enriching aerobic denitrifying functional bacteria. The bioaugmentation mechanism of trehalose on aerobic denitrifying bacteria in high-salt wastewater was investigated in terms of nitrogen removal performance, diversity of bacterial population and abundance of nitrogen removal functional genes. The results showed that the removal rates of NH4+-N, TN and COD in the experimental groups (C40, C120, C360 and C1 080) increased by 10.70%, 32.72%, 27.36%, 19.45% and 8.32%, 28.36%, 22.53%, 17.63% and 12.09%, 31.14%, 25.27%、25.06%, compared to the control group (C0), respectively. The addition of trehalose improved the denitrification efficiency of the bacterial community in high-salt wastewater, and the NH4+-N removal rate increased by up to 32.72% at a concentration of 120 μmol·L-1. High-throughput sequencing analysis revealed that under the high salinity stress, different added trehalose concentrations significantly affected bacterial community structure and abundances. Groups C40 and C120 increased the relative abundances of heterotrophic nitrification-aerobic denitrification (HN-AD) bacteria (including Pseudofulvimonas, Rhodobacteraceae, Paracoccus, Parapusillimonas, and Flavobacterium), while groups C360 and C1 080 favored heterotrophic denitrified bacteria (HDB) (including Enterococcus, Nitrincola, Truepera, and Fusibacter). This demonstrated that trehalose concentration significantly affected the composition and abundance of nitrification and denitrification bacteria in high-salt wastewater. PICRUSt1 suggested that the addition of trehalose effectively increased the denitrification activity of denitrifying microorganism dominated by HN-AD bacteria and HDB, and the relative abundance of nitrification genes (hao) and denitrification genes (nasA, napA and napB) associated with HN-AD bacteria increased in groups C40 and C120, which intensified the HN-AD pathway. However, the relative abundances of denitrification genes (narG, narH and narI) associated with HDB increased in groups C360 and C1 080, indicating that the heterotrophic denitrification pathway was enhanced by the addition of high-level trehalose concentration. In addition, the relative abundance of nitrification and denitrification genes was the highest at the trehalose concentration of 120 μmol·L−1, which maximally accelerated the nitrification and denitrification process of aerobic nitrogen removal bacteria. This study can provide a reference for the application of aerobic nitrogen removal bacteria in the aerobic biological treatment technology in high-salt wastewater. -
食品加工、制药工业、皮革工业、垃圾渗滤液等废水不仅氨氮 (NH4+-N) 浓度高,还含有大量盐分[1-3]。生物法虽然作为一种经济有效的废水处理方法,但是高盐分可抑制微生物酶活,甚至引起细胞质壁分离导致微生物死亡[4-5],不利于微生物对NH4+-N的降解。因此,在对高盐废水的脱氮处理中,通常使用膜分离[6]、蒸发结晶[7]、芬顿[8]和电渗析技术[9]等物化法进行脱盐预处理,这不但使运行成本高、工艺流程长,还会造成二次污染。虽然经物化脱盐处理后的高盐含氮废水可进一步通过生物法脱氮,但去除效果并不稳定[10-11],即使在低盐度环境(<2%),传统硝化、反硝化菌和常规异养菌等常见脱氮微生物的生长代谢及活性也会受到明显抑制[12-13]。具备异养硝化-好氧反硝化(heterotrophic nitrification-aerobic denitrification, HN-AD)功能的好氧脱氮菌在好氧条件下因具备同时硝化反硝化、且无中间产物亚硝态氮 (NO2−-N) 和硝态氮 (NO3−-N) 积累的特质得到广泛关注[14]。不仅如此,部分HN-AD菌属还兼具耐盐性,已被广泛应用于高盐废水处理过程,如不动杆菌属 (Acinetobacter) 、盐单胞菌属 (Halomonas) 、副球菌属 (Paracoccus) [15]等。然而,在工程应用中,许多工业废水的盐度波动大,这使得包括HN-AD菌在内的众多微生物难以快速适应不同的盐度环境,菌群无法实现高效富集,进而影响废水处理效果[16-17]。为了增强微生物对高盐胁迫的抵抗能力,已有研究[18]通过增加盐浓度驯化功能菌属耐盐性能的方式以期获得高去污效率、高富集丰度的耐盐型脱氮功能微生物。然而,在实际应用中却发现,即使经过长期驯化,功能微生物对高盐胁迫的抵抗能力仍较弱[19-20],原因在于功能微生物易流失、难截留导致富集丰度较低,最终影响废水处理效果的稳定性[19, 21-22]。因此,为解决这一问题,亟待研发一种即可提高微生物对高盐胁迫的抵抗能力,又可提高功能菌属生长富集能力的新技术。
微生物在逆境中可通过积累或从环境中吸收相容性溶质 (compatible solutes, CS) 来抵抗外部条件带来的胁迫作用,在不需要细胞内酶的任何特殊适应下即可保护细胞免受不可逆转的损害[23-24]。所以CS在细胞中发挥了重要作用,它不仅不会阻碍正常的细胞代谢,还可以平衡细胞质内的渗透压以增强微生物的活性[23, 25]。在VYRIDES等[26]的研究中,盐度为4%时,假单胞菌株 (Pseudomonas sp strain ADP) 产生的相容性溶质主要为海藻糖;ZHANG等[27]在使用颗粒污泥处理含盐养殖废水的过程中,添加1 000 μmol·L−1的海藻糖,可使产甲烷活性和磷酸酶活性分别提高22%和27%。而海藻糖作为一种价格低、易采购的典型相容性溶质,在高盐废水处理中已被成功应用[28-29]。其作为渗透调节剂易被微生物吸收利用,通过调节细胞渗透压帮助微生物快速适应高盐环境,提升去污能力[26]。目前,已有海藻糖用于高盐废水的厌氧处理系统的相关报道[29-30]。BAI等[30]研究了在序批式反应器中添加海藻糖考察了厌氧氨氧化细菌处理富氮含盐废水的长期脱氮性能,在添加浓度为300 μmol·L−1时,就可使NH4+-N、NO2−-N的去除率分别提高为32.4%和42.2%;杨振琳等[28]通过添加海藻糖强化厌氧化耦合反硝化工艺,在盐度为3.2%的模拟废水中加入250 μmol·L−1的海藻糖,NH4+-N和NO2−-N的去除速率分别提高了81.25%和75%。可见海藻糖对提高长期脱氮效果有显著性作用,但其在高盐废水好氧生物处理中的研究却鲜有报道。盐度显著影响微生物的物理和生化特性,长期作用甚至能够改变整个环境的微生物群落结构[31]。
本研究利用膜曝气生物膜反应器 (membrane aerobic biofilm reactor, MABR) 富集好氧脱氮菌,通过调节外加海藻糖的浓度,采用高通量测序等分析方法,从微生物脱氮性能、群落结构及脱氮功能基因丰度等层面,研究海藻糖浓度梯度对高盐废水中微生物的物种丰度、菌群多样性及脱氮功能基因的影响,旨在为推动好氧脱氮菌在高盐废水好氧生物脱氮处理的实际应用提供理论基础及新技术思路。
1. 材料与方法
1.1 MABR实验装置与运行条件
在实验过程中,建立了5个MABR,工作体积为0.85 L,反应装置见图1。反应器的膜组件由128根中空纤维膜组成,用于空气运输和生物膜附着,中空纤维是用复合聚合物材料制造,只允许气体分子渗透。纤维的外径为300 μm,内径为200 μm,膜孔径为0.1 μm,有效比表面积为1.76 cm2·cm−3。膜组件一端通过流量计和压力表连接到空气泵,另一端连接到底端曝气管形成氧循环,曝气压力设定为10 kPa,进出水均采用蠕动泵完成,水力停留时间为48 h[32]。反应器顶部留有取样口,用于采样和溶解氧 (DO) 监测等操作。
1.2 实验用水及菌液接种
合成的高盐高氮废水由无水CH3COONa (碳源) 、(NH4)2SO4 (氮源) 、NaCl和微量元素组成。进水COD为5 000 mg·L−1,NH4+-N为500 mg·L−1,盐度为2.5%,微量元素为50 mL·L−1。微量元素配方: MgSO4·7H2O 2.0 g·L−1,MnSO4·H2O 0.1 g·L−1,CaCl2 1.5 g·L−1,FeSO4·7H2O 0.1 g·L−1,K2HPO4 5.0 g·L−1。
该研究的接种菌液中主要为产碱杆菌(Alcaligenes)、不动杆菌(Acinetobacter sp.TAC-1)和假单胞菌 (Pseudomonas) 等具备HN-AD功能的好氧脱氮混合菌群。这些细菌主要分离于猪场沼液、垃圾渗滤液和化工废水,可耐受高浓度氨氮。为确保适当的菌液浓度和脱氮活性,首先进行菌液驯化,再分别接种于5个反应器中进行挂膜。挂膜完成后,分别加入不同浓度海藻糖 (0、40、120、360和1 080 μmol·L−1) 进行实验,对应反应器分别为C0、C40、C120、C360和C1 080。
1.3 水质分析方法及生物膜表观特征分析
反应器启动运行阶段的溶氧、盐度及pH值分别采用便携式溶氧仪、海水比重计及pH计进行直接测量。菌液浓度 (OD600 nm) 采用紫外分光光度法测定,进、出水NH4+-N、NO3−-N、NO2−-N、TN和COD的浓度均参照文献中的方法[33]进行检测。
利用扫描电镜 (scanning electron microscope, SEM) 技术表征反应器挂膜阶段的生物膜表观形态特征。将采集的生物膜样品经过固定、PBS清洗、乙醇梯度脱水 (30%、50%、70%、85%和95%质量分数的乙醇洗脱1次,100%乙醇洗脱2次(每次脱水时间为15~20 min)) 、冷冻干燥等步骤完成样品预处理,将制备完成的样品送至武汉铄思百检测技术有限公司并进行SEM观察。
1.4 DNA提取、高通量测序及脱氮功能基因表达
在各反应器稳定运行阶段,采集微生物样品(每个反应器重复采集3次样品,每个样品约0.5 g),将样品短期保存于−80 ℃冰箱中。采用通用检测试剂盒 (MOBIO实验室,美国) 提取样品的总基因组DNA,采用引物338F和806R扩增V3~V4区域的l6S rRNA基因,测序引物序列为338F(5'-ACTCCTACGGGAGGCAGCA-3')和806R(5'-GGACTACHVGGGTWTCTAAT-3')。采用Trimmomatic和Flash软件对测序序列进行拼接,通过Usearch 软件对具有97%相似性的序列进行OTU聚类,经Usearch_global 获得OTU序列丰度统计表,进行后续微生物群落多样性分析;基于京都基因和基因组百科全书(http://www.genome.jp·KEGG)数据库及PICRUSt1软件,对样品中微生物脱氮功能基因进行预测分析。
2. 结果与讨论
2.1 反应器启动及不同海藻糖浓度下的脱氮性能分析
SEM表征结果显示,反应器启动运行20 d,膜丝上明显附着了大量的以杆菌和球菌为主的微生物 (图2) ,说明反应器于20 d内成功完成挂膜启动。该研究反应器菌液挂膜时间较文献报道相关结果[32-34]略有增加,这可能是因为盐度抑制了微生物的生长繁殖,从而导致挂膜周期延长。反应器启动运行阶段的污染物去除情况如图3(a)~图3(e)中第1阶段所示。5个平行反应器的NH4+-N、TN及COD去除率分别为32%~45%、32%~40%和60%~71%,无明显有害中间产物NO2−-N和NO3−-N的积累。启动阶段反应器脱氮性能较低,究其原因主要在于高盐环境对反应器富集的脱氮微生物的生长及活性存在影响。高盐度会使微生物细胞处于一个高渗环境,影响细胞正常生长代谢,进而降低污染物的去除率[35]。
在4个平行反应器中添加不同浓度海藻糖 (40、120、360和1 080 μmol·L−1) ,设定为实验组 (C40、C120、C360和C1 080) ,对照组 (C0) 不添加海藻糖,连续运行80 d结束。添加海藻糖后的反应器运行性能如图3(a)~图3(e)中第2阶段所示。相较于对照组,4个实验组中NH4+-N去除率分别提高了10.70%、32.72%、27.36%、19.45%,TN去除率分别提高了8.32%、28.36%、22.53%、17.63%,COD去除率分别提高了12.09%、31.14%、25.27%、25.06%。这说明添加海藻糖对高盐给微生物带来的高渗胁迫具有缓解作用,提高了系统脱氮性能。C120组的NH4+-N、TN及COD去除率最高,C360组的TN去除率及C1 080组的NH4+-N、TN去除率相较C120组降低,3组中COD去除率无明显变化。可以看出,外源海藻糖浓度与污染物去除率无线性相关性,最适海藻糖浓度为120 μmol·L−1。这主要是因为微生物对海藻糖的最大渗透调节剂有一定的吸收范围[28],当体系内海藻糖浓度超过微生物作为渗透调节剂的吸收浓度范围时,过剩的海藻糖可充当常规异养反硝化菌 (heterotrophic denitrified bacteria, HDB) 的有机碳源[28]。因此,在本研究中,高浓度海藻糖 (360、1 080 μmol·L−1) 条件可能对体系内HDB生长更有利,进而影响好氧脱氮菌群的富集与活性,降低了反应器脱氮性能。
2.2 外源性海藻糖作用下的微生物特性分析
1) 微生物α多样性分析。通过微生物群落分析研究不同海藻糖浓度对反应器中物种丰富度及多样性的影响,结果如表1所示。通过MiSeq Illumina测序分析获得269~303个OTU,覆盖指数均大于99%,说明测序深度包含测序样品中的全部细菌数。Ace指数和Chao指数均为物种丰富度的指标,实验组较对照组有所增加,表明添加海藻糖提升了高盐环境下系统内的物种丰富度。Shannon指数反映微生物多样性特征,其数值同样在实验组中有所增长,说明添加海藻糖丰富了高盐环境下系统内的菌群多样性。C120组的Chao指数、Ace指数以及Shannon指数均最高,说明120 μmol·L−1的海藻糖浓度对增强高盐胁迫下微生物群落的抵抗力稳定性最有利。
表 1 不同海藻糖浓度下微生物Alpha多样性指数Table 1. Alpha diversity indices at different trehalose concentrations反应器 OTUs 覆盖率/% Chao指数 Ace指数 Shannon指数 C0 269 99.82 320.50 314.21 3.06 C40 274 99.82 331.34 320.83 3.24 C120 303 99.82 375.63 370.12 3.83 C360 297 99.78 352.38 367.88 3.66 C1 080 286 99.82 330.13 332.30 3.35 2) 微生物群落组成分析。不同海藻糖浓度下微生物群落组成相似但相对丰度略有差异(图4)。如图4(a)所示,门水平相对丰度排名前3的优势菌门分别为变形菌(Proteobacteria)、拟杆菌(Bacteroidota)和厚壁菌(Firmicutes)。三者的总相对丰度在对照组C0中为82.60%,在各实验组 (C40、C120、C360和C1 080) 中分别提高了3.29%、5.64%、5.49%和3.84%,说明在高盐条件下添加海藻糖对系统中门水平的群落结构具有一定影响。有报道[36-37]指出,Proteobacteria和Bacteroidota在高盐、高氨氮废水处理中常占据主导地位,其包含着多样性最丰富的硝化和反硝化细菌 (如HN-AD菌) ;也有报道[38]指出,可在极端环境进行异养硝化的脱氮微生物 (如肠球菌属Enterococcus) 归属于Firmicutes。该3类菌门中微生物总相对丰度的增加说明添加海藻糖有利于高盐胁迫下脱氮菌群的生长富集。
由属水平相对丰度排名前50的微生物进行层级聚类Heatmap分析结果 (图4(c)) 可知,在C0组中,不动杆菌属 (Acinetobacter) 相对丰度为34.58%,是系统内的优势菌属,Acinetobacter在高氨氮浓度下是硝化和反硝化过程中生长的主要菌群,表现出高效的HN-AD能力[39],且具有耐受高盐、高氨氮和脱氮速率快的优点[40]。而添加海藻糖的4个实验组中,高丰度优势菌属仍为Acinetobacter且相对丰度保持稳定(分别为35.69%、34.95%、34.93%和31.04%)。这说明在高盐环境下,海藻糖对Acinetobacter生长富集的影响较弱,但Acinetobacter保证了高盐系统内的高脱氮效率。在C120组中,假黄褐藻属 (Pseudofulvimonas) 相对丰度明显高于对照组和其他实验组,并且其相丰度变化存在显著性差异 (P <0.05) (图4(b)) 。Pseudofulvimonas相对丰度在低浓度海藻糖时 (40、120 μmol·L−1),随海藻糖浓度的增加而增长,但在高浓度海藻糖时 (360、1 080 μmol·L−1) ,却随海藻糖浓度的增加而减少。Pseudofulvimonas为耐盐型HN-AD菌,是高盐废水处理过程中常见的脱氮功能微生物[41-42];同时,黄杆菌属 (Flavobacterium) [43]、红杆菌属 (Rhodobacteraceae) [44]、假单胞菌属 (Pseudomonas) [16]、副球菌属 (Paracoccus) [45]和产碱菌科 (Parapusillimonas) [46]等耐盐型HN-AD菌在海藻糖浓度作用下的相对丰度变化规律与Pseudofulvimonas一致并同样存在显著性差异 (P<0.05) (见图4(b)) 。Pseudofulvimonas通常与各种氮物质降解过程有关[47];Flavobacterium与生物絮体聚集和颗粒稳定性有关,其丰度增加有利于功能微生物的富集进而保证污染物稳定有效的去除[42, 48];Rhodobacteraceae是海水形成初始生物膜的主要定殖者[44],其具有较强的耐盐性能,能将NH4+-N转化为NO3−-N和NO2−-N,且可在曝气的条件下实现对NO3−-N的高效去除[49-50];Pseudomonas、Paracoccus具有亚硝酸盐还原和有机物降解等多种功能,在有机物去除方面起着关键作用[51-52]。当海藻糖浓度为120 μmol·L−1时,这些HN-AD菌属相对丰度均达到最高。说明低浓度海藻糖促进高盐废水中耐盐型HN-AD菌生长富集,使系统内主要发生异养硝化和好氧反硝化过程,可强化微生物对氮、碳污染物的去除性能。
在C360和C1 080组中,Enterococcus相对丰度由C0中的0.12%分别增至13.88%和21.22% (P <0.05) (图4(b)) 。Enterococcus是一种兼性脱氮菌[53],具有较强的脱氮耐盐性能及高效的产絮凝效果,在生长代谢过程中产生多糖、蛋白质、脂和核酸等成分,可促进水体中菌体细胞凝聚进而减缓功能微生物的流失[54-55]。Enterococcus相对丰度在C360和C1 080组得到明显提升,说明高浓度海藻糖的作用倾向于通过增强高盐条件下微生物凝聚能力,提高功能菌群富集。此外,硝化细菌属 (Nitrincola) 、特吕珀菌属 (Truepera) 和梭菌属 (Fusibacter) 3种HDB的相对丰度在高浓度海藻糖时 (360、1 080 μmol·L−1) 也呈现增长趋势。Nitrincola是一类耐盐碱的厌氧反硝化细菌[15],能够在极端环境中对地球化学元素循环起促进作用[56];Truepera[32]与厌氧反硝化作用密切相关[57],且能够在碱性、中等盐度和高温等多种极端条件下生长,有利于NO向N2O的转换[58-59];Fusibacter是一类耐盐、厌氧的棒杆细菌,可将硫酸盐和元素硫还原成硫化物,还能够广泛利用碳水化合物 (如乙酸、葡萄糖等) 生成CO2和H2等产物[60],且具有脱氮除磷的功能[61]。上述3种菌属均在高浓度氨氮环境的反硝化过程中发挥重要作用,其相对丰度在高海藻糖浓度作用下提高,也证实高浓度海藻糖促进高盐废水中HDB生长富集,使系统内主要发生常规异养反硝化过程。这主要是因为高浓度海藻糖促进Enterococcus相对丰度大幅增加,虽有利于微生物凝聚,但同时也产生大量菌胶团聚集堵塞膜孔,致使生物膜层出现缺氧环境,不利于好氧脱氮微生物的生长,继而引起HDB增殖。虽然HDB相对丰度在高海藻糖浓度下的增长促使C360和C1 080组的NH4+-N和TN去除率相比C0组有所提高,但其相比C40和C120组却呈下降趋势,推测耐盐型HN-AD菌的相对丰度变化对系统脱氮性能的影响相比HDB更为显著和重要,而HDB的相对丰度变化主要影响了系统的脱氮菌群结构。
3) 环境因子相关性分析。通过相关性Heatmap图进一步分析环境因子对系统内属水平群落组成的影响,结果如图5所示。与NH4+-N、COD及TN去除率和海藻糖浓度4个因子均显著关联的只有Pseudomonas的相对丰度,且呈负相关;NH4+-N、COD及TN去除率与Flavobacterium、Parapusillimonas 2种耐盐型HN-AD菌属相对丰度呈正相关,与浮游盐属 (Planktosalinus) 、新巨藻属(Neomegalonema)相对丰度呈负相关。Planktosalinus归属黄杆菌科 (Flavobacteriaceae) ,而Flavobacteriaceae是废水系统发生污泥膨胀的指示微生物[62-63];Neomegalonema同样属于污泥膨胀菌群,其丰度变化用于表征氮磷失衡下废水中的微生物群落健康状况[64-65]。海藻糖浓度与Enterococcus、球毛菌属 (Sphaerochaeta) 和脱硫微杆菌属 (Desulfomicrobium) 相对丰度呈正相关。可以看出,不同菌属相对丰度受到海藻糖浓度、污染物去除率影响的情况存在差异,可证明耐盐型HN-AD菌确为系统内主要脱氮贡献者,显著影响反应器的脱氮性能;以Enterococcus为代表的传统脱氮菌和其他功能微生物的相对丰度变化规律则主要反映了海藻糖浓度梯度对高盐废水中脱氮菌群组成的差异影响。
2.3 脱氮功能基因预测分析
采用PICRUSt1对功能基因丰度进行分析,其相对丰度变化结果见图6(a)。该研究获得与硝化过程相关的羟胺氧化酶基因hao,是NH2OH氧化生成NO2−-N过程的关键酶基因[66],其在实验组 (C40、C120、C360和C1 080) 中相对丰度相较对照组 (C0) 分别提高了4.96倍、34.70倍、32.45倍和29.06倍,说明添加海藻糖明显加快系统内微生物的硝化进程。同时,还获得8个与反硝化过程相关的功能基因,包括硝酸盐还原酶(nasA、nasB、narG、narH、narI、napA、napB)、亚硝酸盐还原酶(nirK)、一氧化氮氧化酶(norB、norC)和一氧化二氮还原酶(nosZ)。4个实验组中反硝化功能基因总相对丰度较对照组分别提高了1.31倍、2.86倍、1.81倍和1.75倍,说明添加海藻糖同样促进了系统内微生物的反硝化进程。
硝酸还原酶催化细菌体内NO3−-N还原生成NO2−-N,是实现反硝化过程的关键酶之一。nasA是一种同化型硝酸盐还原酶基因,存在于细菌的细胞质中[67],也常见于HN-AD菌[68]。其在实验组中的相对丰度值相较对照组均有提高,低浓度海藻糖时 (40、120 μmol·L−1) ,nasA基因相对丰度随海藻糖浓度的增加而增长,于海藻糖浓度为120 μmol·L−1时达到最高值;但在高浓度海藻糖时 (360、1 080 μmol·L−1) ,其随海藻糖浓度增加出现降低。同样,异化型硝酸盐还原酶NAP的基因napA和napB相对丰度变化规律与nasA一致。NAP表达在好氧条件下占主导地位,常存于HN-AD菌中[69]。另一种异化型硝酸还原酶NAR的基因narG、narH和narI相对丰度却在海藻糖浓度为360 μmol·L−1和1 080 μmol·L−1时增加,而NAR表达在缺氧条件下占主导地位,常见于HDB[70]。以上结果说明,在高盐废水添加海藻糖可诱导增强以HN-AD菌和HDB为主的脱氮菌群的反硝化活性。低浓度海藻糖倾向于强化系统的异养硝化-好氧反硝化途径,而高浓度海藻糖则偏向于促进系统的异养反硝化途径 (图6(b)) 。
亚硝酸盐还原酶NIR是反硝化过程的另一类关键酶,驱动NO2−-N还原生成气体产物NO,存在于细胞膜外生物细胞质中,其基因有nirS和nirK 2种类型[71]。海藻糖的加入提高了nirK的相对丰度,且在海藻糖浓度为120 μmol·L−1时达到最高值,推进了系统的亚硝酸盐还原进程。NO还原酶基因 (norB和norC) 和nosZ分别是催化NO还原生成N2O、N2O进一步还原生成最终产物N2的关键酶基因。在污水处理中,反硝化细菌体内nosZ活性的降低或不具有nosZ是导致反硝化过程中温室气体N2O大量积累的主要因素[72]。该研究nosZ的相对丰度随着海藻糖浓度的增加而升高,说明添加海藻糖对控制好氧脱氮过程的N2O产生与积累具有一定作用。以上基因预测结果也进一步证明,海藻糖浓度为120 μmol·L−1时,反应器的NH4+-N、TN去除率最高 (图2) 。这主要是因为此海藻糖浓度条件激发HN-AD菌的硝化和反硝化进程达到最快,极大程度加速了NH4+-N氧化及NO3−-N和NO2−-N的还原,最终使反应器脱氮性能达到最优。
3. 结论
1) 添加海藻糖的实验组 (40、120、360和1 080 μmol·L−1) 中NH4+-N、TN和COD去除率相较对照组 (0 μmol·L−1) 显著提高,最适海藻糖浓度为120 μmol·L−1。外加海藻糖可优化高盐条件下的反应器脱氮性能。
2) 海藻糖浓度对脱氮菌群的相对丰度具有显著影响。低浓度海藻糖 (40、120 μmol·L−1) 有利于耐盐型HN-AD菌相对丰度的提高,而高浓度海藻糖 (360、1 080 μmol·L−1) 更倾向于促进HDB的富集;耐盐型HN-AD菌是系统的主要脱氮贡献者,其相对丰度变化与污染物去除率之间存在显著相关性,传统脱氮菌相对丰度变化则与高盐废水中脱氮菌群组成差异有关。耐盐型HN-AD菌对调控高盐条件下的反应器脱氮性能起着重要影响。
3) 海藻糖可有效提高以HN-AD菌和HDB为主的脱氮菌群的硝化和反硝化活性。在低浓度海藻糖 (40、120 μmol·L−1) 作用下,与HN-AD菌关联的硝化、反硝化基因(hao、nasA、napA和napB)丰度随海藻糖浓度的增加而增长,系统的异养硝化-好氧反硝化途径得到强化;在高浓度海藻糖 (360、1 080 μmol·L−1) 作用下,与HDB关联的反硝化基因 (narG、narH和narI) 相对丰度明显增加,系统的异养反硝化途径得到增强。
-
表 1 不同海藻糖浓度下微生物Alpha多样性指数
Table 1. Alpha diversity indices at different trehalose concentrations
反应器 OTUs 覆盖率/% Chao指数 Ace指数 Shannon指数 C0 269 99.82 320.50 314.21 3.06 C40 274 99.82 331.34 320.83 3.24 C120 303 99.82 375.63 370.12 3.83 C360 297 99.78 352.38 367.88 3.66 C1 080 286 99.82 330.13 332.30 3.35 -
[1] 亓金鹏, 肖小兰, 张瑞娜, 等. AnMBR 处理高盐榨菜废水的运行效能及膜污染特性[J]. 环境工程学报, 2021, 15(2): 553-562. doi: 10.12030/j.cjee.202004134 [2] 冯雅丽, 张茜, 李浩然, 等. 铁炭微电解预处理高浓度高盐制药废水[J]. 环境工程学报, 2012, 6(11): 3855-3860. [3] ZHAO Y, ZHUANG X, AHMAD S, et al. Biotreatment of high-salinity wastewater: current methods and future directions[J]. World Journal of Microbiology and Biotechnology, 2020, 36(3): 1-11. [4] 张彦灼, 李军, 陈光辉, 等. NaCl 对好氧颗粒污泥短程硝化反硝化的影响[J]. 环境科学研究, 2015, 28(5): 823-830. [5] LI P, CHEN Q, DONG H, et al. Effect of applying potentials on anaerobic digestion of high salinity organic wastewater[J]. Science of the Total Environment, 2022, 822: 153416. doi: 10.1016/j.scitotenv.2022.153416 [6] DASTGHEIB S A, SALIH H H. Treatment of highly saline brines by supercritical precipitation followed by supercritical membrane separation[J]. Industrial & Engineering Chemistry Research, 2019, 58(8): 3370-3376. [7] SHI J, HUANG W, HAN H, et al. Review on treatment technology of salt wastewater in coal chemical industry of China[J]. Desalination, 2020, 493: 114640. doi: 10.1016/j.desal.2020.114640 [8] ZHANG W, YANG X, WANG D. Complete removal of organic contaminants from hypersaline wastewater by the integrated process of powdered activated carbon adsorption and thermal Fenton oxidation[J]. Industrial & Engineering Chemistry Research, 2013, 52(16): 5765-5771. [9] 张玉浩, 陈彦洁, 付国楷. 电流强度对高盐废水混合生物阴极MFC脱氮及产电的影响[J]. 环境工程学报, 2021, 15(7): 2436-2449. doi: 10.12030/j.cjee.202011122 [10] SRIVASTAVA A, PARIDA V K, MAJUMDER A, et al. Treatment of saline wastewater using physicochemical, biological, and hybrid processes: Insights into inhibition mechanisms, treatment efficiencies and performance enhancement[J]. Journal of Environmental Chemical Engineering, 2021, 9(4): 105775. doi: 10.1016/j.jece.2021.105775 [11] 王忠泉. 微电解/曝气生物流化床工艺处理高盐废水的研究[J]. 中国给水排水, 2019, 35(7): 5. doi: 10.19853/j.zgjsps.1000-4602.2019.07.016 [12] HE H, CHEN Y, LI X, et al. Influence of salinity on microorganisms in activated sludge processes: A review[J]. International Biodeterioration & Biodegradation, 2017, 119: 520-527. [13] HUANG J L, CUI Y W, YAN J L, et al. Occurrence of heterotrophic nitrification-aerobic denitrification induced by decreasing salinity in a halophilic AGS SBR treating hypersaline wastewater[J]. Chemical Engineering Journal, 2022, 431: 134133. doi: 10.1016/j.cej.2021.134133 [14] 袁建华, 赵天涛, 彭绪亚. 极端条件下异养硝化-好氧反硝化菌脱氮的研究进展[J]. 生物工程学报, 2019, 35(6): 942-955. doi: 10.13345/j.cjb.180427 [15] PAN Z, ZHOU J, LIN Z, et al. Effects of COD/TN ratio on nitrogen removal efficiency, microbial community for high saline wastewater treatment based on heterotrophic nitrification-aerobic denitrification process[J]. Bioresource Technology, 2020, 301: 122726. doi: 10.1016/j.biortech.2019.122726 [16] FU G, HAN J, YU T, et al. The structure of denitrifying microbial communities in constructed mangrove wetlands in response to fluctuating salinities[J]. Journal of Environmental Management, 2019, 238: 1-9. [17] JIN R C, YANG G F, YU J J, et al. The inhibition of the Anammox process: A review[J]. Chemical engineering journal, 2012, 197: 67-79. doi: 10.1016/j.cej.2012.05.014 [18] LIN L, PRATT S, RATTIER M, et al. Individual and combined effect of salinity and nitrite on freshwater Anammox bacteria (FAB)[J]. Water Research, 2020, 169: 114931. doi: 10.1016/j.watres.2019.114931 [19] FANG F, YANG M M, WANG H, et al. Effect of high salinity in wastewater on surface properties of anammox granular sludge[J]. Chemosphere, 2018, 210: 366-375. doi: 10.1016/j.chemosphere.2018.07.038 [20] MENG Y, YIN C, ZHOU Z, et al. Increased salinity triggers significant changes in the functional proteins of ANAMMOX bacteria within a biofilm community[J]. Chemosphere, 2018, 207: 655-664. doi: 10.1016/j.chemosphere.2018.05.076 [21] PATIL P K, BASKARAN V, VINAY T N, et al. Abundance, community structure and diversity of nitrifying bacterial enrichments from low and high saline brackishwater environments[J]. Letters in Applied Microbiology, 2021, 73(1): 96-106. doi: 10.1111/lam.13480 [22] 殷豪帅, 黄开, 王卿卿, 等. 高盐度环境下某污水处理厂AAO生化系统微生物群落变化分析[J]. 环境工程, 2021, 39(3): 68-74. doi: 10.13205/j.hjgc.202103010 [23] VYRIDES I, STUCKEY D C. Adaptation of anaerobic biomass to saline conditions: Role of compatible solutes and extracellular polysaccharides[J]. Enzyme and Microbial Technology, 2009, 44(1): 46-51. doi: 10.1016/j.enzmictec.2008.09.008 [24] CYPLIK P, PIOTROWSKA-CYPLIK A, MARECIK R, et al. Biological denitrification of brine: The effect of compatible solutes on enzyme activities and fatty acid degradation[J]. Biodegradation, 2012, 23(5): 663-672. doi: 10.1007/s10532-012-9542-0 [25] ZHANG Z Z, JI Y X, CHENG Y F, et al. Increased salinity improves the thermotolerance of mesophilic anammox consortia[J]. Science of the Total Environment, 2018, 644: 710-716. doi: 10.1016/j.scitotenv.2018.07.027 [26] VYRIDES I, STUCKEY D C. Compatible solute addition to biological systems treating waste/wastewater to counteract osmotic and other environmental stresses: A review[J]. Critical Reviews in Biotechnology, 2017, 37(7): 865-879. doi: 10.1080/07388551.2016.1266460 [27] ZHANG X, FERREIRA R B, HU J, et al. Improving methane production and phosphorus release in anaerobic digestion of particulate saline sludge from a brackish aquaculture recirculation system[J]. Bioresource technology, 2014, 162: 384-388. doi: 10.1016/j.biortech.2014.04.005 [28] 杨振琳, 于德爽, 李津, 等. 海藻糖强化厌氧氨氧化耦合反硝化工艺处理高盐废水的脱氮除碳效能[J]. 环境科学, 2018, 39(10): 4612-4620. doi: 10.13227/j.hjkx.201803218 [29] VYRIDES I, SANTOS H, MINGOTE A, et al. Are compatible solutes compatible with biological treatment of saline wastewater? Batch and continuous studies using submerged anaerobic membrane bioreactors (SAMBRs)[J]. Environmental Science & Technology, 2010, 44(19): 7437-7442. [30] BAI L, LI J, LI R, et al. Long‐term nitrogen removal performance and kinetics of anaerobic ammonia oxidation bacteria treating nitrogen‐rich saline wastewater with trehalose addition[J]. Water Environment Research, 2019, 91(11): 1518-1525. doi: 10.1002/wer.1152 [31] MA C, JIN R C, YANG G F, et al. Impacts of transient salinity shock loads on Anammox process performance[J]. Bioresource Technology, 2012, 112: 124-130. doi: 10.1016/j.biortech.2012.02.122 [32] 康宝文, 肖芃颖, 周靖, 等. 生物膜层DO浓度对MABR中异养硝化-好氧反硝化的影响[J]. 环境科学研究, 2021, 34(10): 2397-2404. doi: 10.13198/j.issn.1001-6929.2021.05.19 [33] 国家环境保护总局. 水和废水监测分析方法[M]. 4版. 北京: 中国环境科学出版社, 2012. [34] 罗晓静, 肖芃颖, 康宝文, 等. MABR快速富集HN-AD菌强化处理高氨氮废水[J]. 环境工程, 2020, 38(9): 106-112. doi: 10.13205/j.hjgc.202009018 [35] HOU T, ZHAO J, LEI Z, et al. Addition of air-nanobubble water to mitigate the inhibition of high salinity on co-production of hydrogen and methane from two-stage anaerobic digestion of food waste[J]. Journal of Cleaner Production, 2021, 314: 127942. doi: 10.1016/j.jclepro.2021.127942 [36] DEVPURA N, JAIN K, PATEL A, et al. Metabolic potential and taxonomic assessment of bacterial community of an environment to chronic industrial discharge[J]. International Biodeterioration & Biodegradation, 2017, 123: 216-227. [37] HE Q, SONG Q, ZHANG S, et al. Simultaneous nitrification, denitrification and phosphorus removal in an aerobic granular sequencing batch reactor with mixed carbon sources: Reactor performance, extracellular polymeric substances and microbial successions[J]. Chemical Engineering Journal, 2018, 331: 841-849. doi: 10.1016/j.cej.2017.09.060 [38] HE T, GUAN W, LUAN Z, et al. Spatiotemporal variation of bacterial and archaeal communities in a pilot-scale constructed wetland for surface water treatment[J]. Applied Microbiology and Biotechnology, 2016, 100(3): 1479-1488. doi: 10.1007/s00253-015-7072-5 [39] MA W, HAN Y, MA W, et al. Enhanced nitrogen removal from coal gasification wastewater by simultaneous nitrification and denitrification (SND) in an oxygen-limited aeration sequencing batch biofilm reactor[J]. Bioresource Technology, 2017, 244: 84-91. doi: 10.1016/j.biortech.2017.07.083 [40] ALI A, WU Z, LI M, et al. Carbon to nitrogen ratios influence the removal performance of calcium, fluoride, and nitrate by Acinetobacter H12 in a quartz sand-filled biofilm reactor[J]. Bioresource Technology, 2021, 333: 125154. doi: 10.1016/j.biortech.2021.125154 [41] SONG X, YU D, QIU Y, et al. Unexpected phosphorous removal in a Candidatus_Competibacter and Defluviicoccus dominated reactor[J]. Bioresource Technology, 2022, 345: 126540. doi: 10.1016/j.biortech.2021.126540 [42] WANG H, GUO L, REN X, et al. Enhanced aerobic granular sludge by static magnetic field to treat saline wastewater via simultaneous partial nitrification and denitrification (SPND) process[J]. Bioresource Technology, 2022, 350: 126891. doi: 10.1016/j.biortech.2022.126891 [43] MENG F, HUANG W, LIU D, et al. Application of aerobic granules-continuous flow reactor for saline wastewater treatment: Granular stability, lipid production and symbiotic relationship between bacteria and algae[J]. Bioresource Technology, 2020, 295: 122291. doi: 10.1016/j.biortech.2019.122291 [44] ELIFANTZ H, HORN G, AYON M, et al. Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater[J]. FEMS Microbiology Ecology, 2013, 85(2): 348-357. doi: 10.1111/1574-6941.12122 [45] SILVA L C F, LIMA H S, SARTORATTO A, et al. Effect of salinity in heterotrophic nitrification/aerobic denitrification performed by acclimated microbiota from oil-produced water biological treatment system[J]. International Biodeterioration & Biodegradation, 2018, 130: 1-7. [46] LI J, QI M, LAI Q, et al. Pusillimonas maritima sp. nov., isolated from surface seawater[J]. International Journal of Systematic and Evolutionary Microbiology, 2020, 70(5): 3483-3490. doi: 10.1099/ijsem.0.004202 [47] ZAINUDIN M H, MUSTAPHA N A, MAEDA T, et al. Biochar enhanced the nitrifying and denitrifying bacterial communities during the composting of poultry manure and rice straw[J]. Waste Management, 2020, 106: 240-249. doi: 10.1016/j.wasman.2020.03.029 [48] XIAO P Y, ZHOU J, LUO X, et al. Enhanced nitrogen removal from high-strength ammonium wastewater by improving heterotrophic nitrification-aerobic denitrification process: insight into the influence of dissolved oxygen in the outer layer of the biofilm[J]. Journal of Cleaner Production, 2021, 297: 126658. doi: 10.1016/j.jclepro.2021.126658 [49] ZHANG Q, CHEN X, ZHANG Z, et al. Performance and microbial ecology of a novel moving bed biofilm reactor process inoculated with heterotrophic nitrification-aerobic denitrification bacteria for high ammonia nitrogen wastewater treatment[J]. Bioresource Technology, 2020, 315: 123813. doi: 10.1016/j.biortech.2020.123813 [50] HUANG X, DONG W, WANG H, et al. Biological nutrient removal and molecular biological characteristics in an anaerobic-multistage anaerobic/oxic (A-MAO) process to treat municipal wastewater[J]. Bioresource Technology, 2017, 241: 969-978. doi: 10.1016/j.biortech.2017.05.161 [51] XU S, ZHANG F, JIANG Y, et al. Characterization of a new heterotrophic nitrification bacterium Pseudomonas sp. strain JQ170 and functional identification of nap gene in nitrite production[J]. Science of the Total Environment, 2022, 806: 150556. doi: 10.1016/j.scitotenv.2021.150556 [52] ZHANG Z, GUO Y, GUO L, et al. Elucidating salinity adaptation and shock loading on denitrification performance: focusing on microbial community shift and carbon source evaluation[J]. Bioresource Technology, 2020, 305: 123030. doi: 10.1016/j.biortech.2020.123030 [53] HEYLEN K, VANPARYS B, WITTEBOLLE L, et al. Cultivation of denitrifying bacteria: Optimization of isolation conditions and diversity study[J]. Applied and Environmental Microbiology, 2006, 72(4): 2637-2643. doi: 10.1128/AEM.72.4.2637-2643.2006 [54] YOKOI H, ARATAKE T, HIROSE J, et al. Simultaneous production of hydrogen and bioflocculant by Enterobacter sp. BY-29[J]. World Journal of Microbiology and Biotechnology, 2001, 17(6): 609-613. doi: 10.1023/A:1012463508364 [55] 刘洪艳, 王红玉. 潮间带污泥产絮凝细菌W17分离与絮凝性质分析[J]. 环境科学与技术, 2015, 38(8): 127-130. [56] VALDÉS N, RIVERA-ARAYA J, BIJMAN J, et al. Draft genome sequence of Nitrincola sp. strain A-D6, an arsenic-resistant gammaproteobacterium isolated from a salt flat[J]. Genome Announcements, 2014, 2(6): e01144-14. [57] 张小姗, 温春晓, 何宁. 曝气生物滤池处理城镇污水厂尾水的强化脱氮及微生物群落特征分析[J]. 生态科学, 2021, 40(6): 48. doi: 10.14108/j.cnki.1008-8873.2021.06.006 [58] IVANOVA N, ROHDE C, MUNK C, et al. Complete genome sequence of Truepera radiovictrix type strain (RQ-24T)[J]. Standards in Genomic Sciences, 2011, 4(1): 91-99. doi: 10.4056/sigs.1563919 [59] LIU C, YAN J, HUANG Q, et al. The addition of sawdust reduced the emission of nitrous oxide in pig manure composting by altering the bacterial community structure and functions[J]. Environmental Science and Pollution Research, 2022, 29(3): 3733-3742. doi: 10.1007/s11356-021-15786-2 [60] SMII L, HANIA W B, CAYOL J L, et al. Fusibacter bizertensis sp. nov., isolated from a corroded kerosene storage tank[J]. International Journal of Systematic and Evolutionary Microbiology, 2015, 65: 117-121. doi: 10.1099/ijs.0.066183-0 [61] 郭海娟, 顾一宁, 马放, 等. 好氧颗粒污泥处理市政污水性能与微生物特性研究[J]. 环境科学学报, 2020, 40(10): 3688-3695. doi: 10.13671/j.hjkxxb.2020.0404 [62] ZHONG Z P, LIU Y, WANG F, et al. Planktosalinus lacus gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from a salt lake[J]. International Journal of Systematic and Evolutionary Microbiology, 2016, 66(5): 2084-2089. doi: 10.1099/ijsem.0.000997 [63] 端正花, 潘留明, 陈晓欧, 等. 低温下活性污泥膨胀的微生物群落结构研究[J]. 环境科学, 2016, 37(3): 1070-1074. doi: 10.13227/j.hjkx.2016.03.036 [64] HERBST F A, GONÇALVES S C L, BEHR T, et al. Proteogenomic refinement of the Neomegalonema perideroedesT genome annotation[J]. Proteomics, 2019, 19(9): 1800330. doi: 10.1002/pmic.201800330 [65] 贺雪濛, 丁丽丽, 张璐璐, 等. 氮磷失衡下膨胀污泥性能及膨胀菌群落结构变化[J]. 环境科学, 2018, 39(4): 1782-1793. doi: 10.13227/j.hjkx.201708094 [66] KUYPERS M M M, MARCHANT H K, KARTAL B. The microbial nitrogen-cycling network[J]. Nature Reviews Microbiology, 2018, 16(5): 263-276. doi: 10.1038/nrmicro.2018.9 [67] LUQUE-ALMAGRO V M, GATES A J, MORENO-VIVIÁN C, et al. Bacterial nitrate assimilation: Gene distribution and regulation[J]. Biochemical Society Transactions, 2011, 39(6): 1838-1843. doi: 10.1042/BST20110688 [68] TAN X, YANG Y L, LIU Y W, et al. Quantitative ecology associations between heterotrophic nitrification-aerobic denitrification, nitrogen-metabolism genes, and key bacteria in a tidal flow constructed wetland[J]. Bioresource Technology, 2021, 337: 125449. doi: 10.1016/j.biortech.2021.125449 [69] ZHU L, DING W, FENG L, et al. Isolation of aerobic denitrifiers and characterization for their potential application in the bioremediation of oligotrophic ecosystem[J]. Bioresource Technology, 2012, 108: 1-7. doi: 10.1016/j.biortech.2011.12.033 [70] NAN X, MA B, QIAN W, et al. Achieving nitritation by treating sludge with free nitrous acid: The effect of starvation[J]. Bioresource Technology, 2019, 271: 159-165. doi: 10.1016/j.biortech.2018.09.113 [71] YU Q, ZHOU R, WANG Y, et al. Corpse decomposition increases nitrogen pollution and alters the succession of nirK-type denitrifying communities in different water types[J]. Science of the Total Environment, 2020, 747: 141472. doi: 10.1016/j.scitotenv.2020.141472 [72] ZOU X, ZHOU Y, GAO M, et al. Effective N2O emission control during the nitritation/denitritation treatment of ammonia rich wastewater[J]. Journal of Environmental Chemical Engineering, 2022, 10(2): 107234. doi: 10.1016/j.jece.2022.107234 -




 下载:
下载: