-
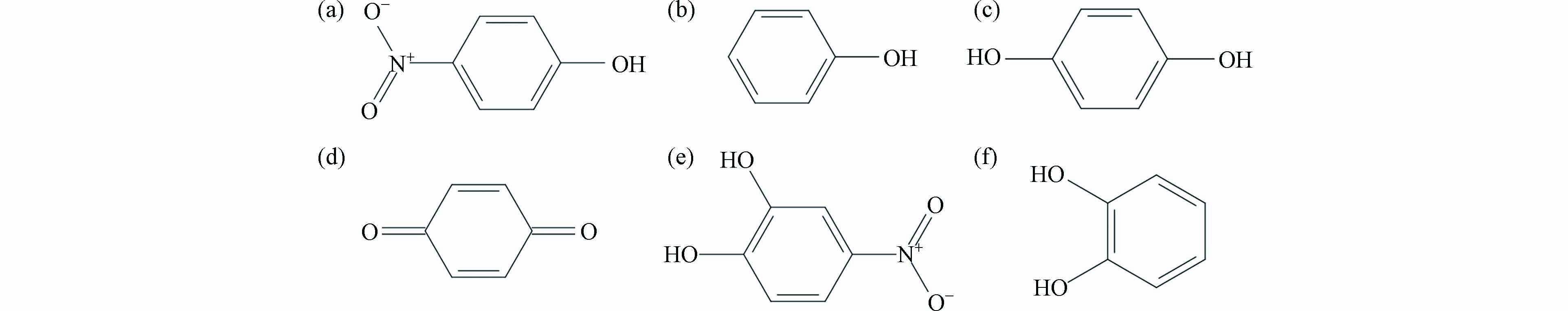
对硝基苯酚(p-nitrophenol,p-NP)是一种重要的有机合成材料,同时也是典型内分泌干扰物和难降解有机污染物,在自然环境中半衰期较长,对水生生物、水体环境和人体健康均有不利影响[1 − 3]. 美国环境保护署将p-NP列为水体中的优先控制污染物,同时p-NP也是我国饮用水水质标准中的污染物控制指标之一[4 − 6].
p-NP具有雌激素活性和抗雄激素活性, 研究表明p-NP可能分别与雌激素受体和雄激素受体相互作用[7]. Li等[8]通过体内实验发现每天向未成熟雌性大鼠注射p-NP会导致子宫重量显著增加,而向未成熟雄性大鼠施用p-NP则会导致生长依赖雄激素的副性腺重量减少. 而Zhang等[9]研究发现,经p-NP治疗的未成熟雄性大鼠的血清雄激素浓度和雄激素受体表达水平均上调,这表明p-NP抗雄激素活性的分子机制是复杂的. 雄激素受体活性通过直接蛋白-蛋白相互作用调节,应激诱导的热激蛋白(heat shock protein 90,HSP)与伴侣蛋白FK506结合蛋白(FK506 binding protein,FKBP5)通过形成FKBP5-Hsp90超复合物来调节雄激素受体活性[10]. 尽管p-NP在体内具有抗雄激素作用,但p-NP抑制雄激素受体信号的分子机制仍不十分清楚. Wu等[11]来用微摩尔亲和力(micromolar affinity)表征了p-NP与FKBP51的结合,实验和分子动力学模拟结果发现p-NP稳定地结合在FK1区域,p-NP结合的热点残基主要是疏水性官能团,p-NP可能通过下调雄激素受体活性激活下游基因的表达水平抑制人类前列腺癌细胞的雄激素依赖性生长,研究结果可以为评估p-NP的潜在健康影响提供新的指导.
近年来,分子对接技术作为计算生物学重要组成部分,在药物研发、生物修复以及计算毒理学等领域起到重要作用[12 − 13]. Ding等[14]利用分子对接技术研究了对羟基苯甲酸酯类物质侧链官能团对其抗雄激素活性的影响,结果表明侧链官能团的种类和长度对抗雄激素活性存在显著影响,同时对羟基苯甲酸酯类物质抗雄激素活性与结合能呈线性关系. Ng等[15]为了研究全氟化合物的生物累积性,利用分子对接技术预测其结合强度和生物半衰期,结果表明全氟辛烷磺酸能够成功对接到人血清白蛋白上且偏差小于2 Å,为新污染物的环境风险管控提供新的思路. 江文婷等[16]利用分子对接技术研究抗冻多肽与海鲈鱼肌球蛋白重链的作用位点及可能的作用机制,结果表明肽段GPR和GPAGGK能够通过碳氢键、氢键和范德华力等分子作用力对海鲈鱼肌球蛋白的结构及聚集行为产生影响,阻碍蛋白侧链聚集、结构改变和冰晶的位移等.
有关研究已经对p-NP在环境中的降解路径和降解产物进行了研究,然而环境中p-NP降解产物(p-NP degradation products,p-NP-DPs)与FKBP5相互作用及其分子水平机制的研究尚未开展. 赵子晗[17]利用紫外全谱扫描和液质联用测定p-NP-DPs,主要有对苯二酚、邻苯二酚、4-硝基邻苯二酚、1,2,4-苯三酚、对苯醌、苯酚等6种物质. Yang等[18]发现环境中p-NP-DPs主要包括对苯二酚、邻苯二酚、4-硝基邻苯二酚、对苯醌、苯酚以及小分子有机酸. p-NP-DPs与和FKBP5相互作用机理尚不清晰,而p-NP-DPs与FKBP5相互作用性能和机理对于理解和防控环境中p-NP对人体的健康风险十分重要. 小分子污染物的降解产物由于具有污染物类似的结构而具有相似的环境效应,污染物与受体蛋白的相互作用受到官能团和分子量等理化性质的影响[19 − 22]. Anzelle等[23]通过研究亚甲基蓝结构类似物对单胺氧化酶抑制作用的影响,结果发现亚甲基蓝、Azure B和ethylthioninium chloride(ETC)对单胺氧化酶A(monoamine oxidase A,MAOA)和单胺氧化酶B(monoamine oxidase B,MAOB)的半数抑制浓度(half maximal inhibitory concentration,IC50)分别为0.07 μmol·L−1(亚甲基蓝-MAOA)、0.01 μmol·L−1(Azure B-MAOA)和0.51 μmol·L−1(ETC-MAOA),4.47 μmol·L−1(亚甲基蓝-MAOB)、0.97 μmol·L−1(Azure B-MAOB)和0.59 μmol·L−1(ETC-MAOB). 袁霞等[24]通过HRMS Orbitrap和GC-MS鉴定出UV/氯高级氧化工艺降解卡马西平的10种降解产物,并利用发光细菌毒性实验和ECOSAR预测卡马西平降解产物的毒性,结果表明卡马西平在UV/氯高级氧化工艺中产生更高毒性的中间产物,对水质安全和污染物毒性管控造成潜在风险.
环境中p-NP-DPs与FKBP5结合形成p-NP-DPs-FKBP5加合物是产生雌激素和抗雄激素活性的主要机制[8 − 10],而p-NP-DPs与FKBP5结合的分子水平机制亟需阐述清楚. 基于此,本文采用通过分子对接技术模拟计算p-NP-DPs与FKBP5相互作用的结合能和结合面积. 在此基础上,通过Spearman相关性和Pearson相关性分析探讨p-NP-DPs理化性质对p-NP-DPs与FKBP5结合的影响,剖析影响p-NP-DPs与FKBP5结合的关键因子,从分子水平阐述p-NP-DPs与FKBP5相互作用的本质. 研究结果有助于增进人们对水环境中p-NP-DPs新污染物分子水平健康效应和环境风险的认识和理解.
-
选择用于分子对接研究的p-NP-DPs为p-NP、苯酚、对苯二酚、对苯醌、4-硝基邻苯二酚和邻苯二酚,化学结构式及其理化性质分别见图1和表1. 根据p-NP-DPs的CAS号,通过PubChem网站(https://pubchem.ncbi.nlm.nih.gov/)下载小分子三维结构,利用分子可视化软件分别对配体小分子进行电荷、质子化状态确认. 通过AutoDock 4.2软件Minimized模块对小分子进行能量最优化、加氢、加电荷及完善结构处理实现小分子的结构修饰,最后将p-NP-DPs的三维结构分别保存为pdb格式文件作为配体结构.
FKBP5原始结构(PDB: 5OMP)从国际标准蛋白质数据库(http://www.rcsb.org/)下载[25]. 通过分子可视化软件对FKBP5进行去除水分子和不需要的杂原子等操作处理,然后通过AutoDock 4.2软件Minimized模块将目的蛋白结构进行能量最优化,同时进行加氢、加电荷及补充大分子的氨基酸残缺结构等完善蛋白结构,最后将FKBP5的晶体结构保存为pdb格式作为受体结构.
打开AutoDock 4.2软件,导入受体结构,确定分子对接口袋区;导入配体结构,通过Dock模块进行分子对接. 通过给出配体与受体结合时氢键、疏水作用、结合面积、能量变化、药效团等参数,对p-NP-DPs与FKBP5相互作用及其分子机制进行解析.
-
采用SPSS 25.0软件(IBM, Armonk,NY,美国)对实验结果进行统计分析. 采用Pearson相关性和Spearman相关性对p-NP-DPs与FKBP5相互作用参数和p-NP-DPs理化性质之间的相关性进行分析,P<0.05被认为具有统计学意义. 根据公式(1)—(4)计算 Pearson 相关性:
式中SX i 和SY j是样本Xi和样本Yj的标准差,Cov(Xi,Yj)是样本Xi和样本Yj的协方差,m表示样本中个体的序号,n表示样本的容量. 样本Xi代表p-NP-DPS的11种理化性质,当i=1, 2…11时分别对应分子量、熔点、沸点、闪点、密度、lgSw、亨利常数、lgP、拓扑极表面积、氢键供体数量和氢键受体数量. 样本Yj代表p-NP-DPS与FKBP5相互作用强度,当j=1, 2时分别对应结合能绝对值和结合面积.
根据公式(5)计算Spearman相关性:
式中,dm是样本Xi和Yj中的第m个体的等级差,n表示样本的容量. 样本Xi代表p-NP和p-NP-DPS的11种理化性质,当i=1, 2…11时分别对应分子量、熔点、沸点、闪点、密度、lgSw、亨利常数、lgP、拓扑极表面积、氢键供体数量和氢键受体数量. 样本Yj代表p-NP-DPS与FKBP5相互作用强度,当j=1, 2时分别对应结合能绝对值和结合面积.
-
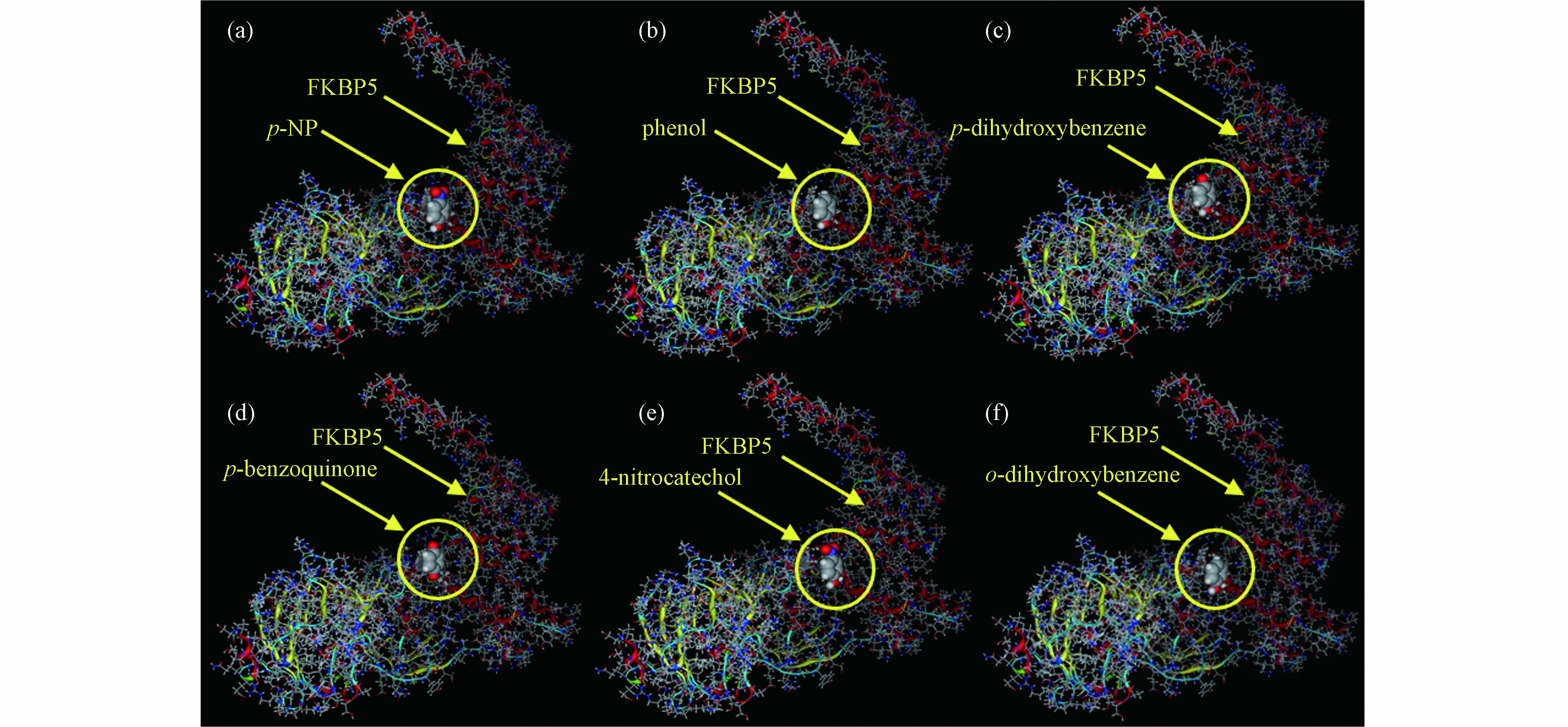
分子对接是目前计算化学中一种重要的研究方法,可用于p-NP-DPs与FKBP5相互作用结合位点和相互作用的研究[26 − 27]. p-NP-DPs与FKBP5分子对接后的最优位置如图2所示. 由图2(a)可知,p-NP与FKBP5相互作用的关键氨基酸为Glu227、Ala228、Lys254、Glu255、Gly282、Met285、Gln286和Trp297. p-NP与FKBP5的相互作用包括—OH→Gly282(氢键)、—OH→Glu227(氢键)、—NO2←Met285(氢键)、—OH→Gln286(氢键)、—OH→Lys254(氢键)、苯环-Trp297(π-π/范德华力相互作用)、苯环-Glu227(范德华力相互作用)、—OH→Glu255(氢键)和苯环-Ala228(范德华力相互作用). 受体和供体相互作用的强度由结合能和结合面积决定,p-NP与FKBP5相互作用的结合能和结合面积分别为−3.76 kcal·mol−1和200 Å2.
如图2(b)所示,苯酚与FKBP5相互作用的关键氨基酸为Pro221、Glu227、Lys254、Glu255、Lys283、Met285、Gln286和Trp297. 苯酚与FKBP5的相互作用包括—OH→Gln286(氢键)、—OH→Pro221(氢键)、—OH→Lys254(氢键)、苯环-Trp297(π-π/范德华力相互作用)、—OH→Glu227(氢键)、苯环-Met285(范德华力相互作用)、苯环-Lys283(范德华力相互作用)、苯环-Glu227(范德华力相互作用)和—OH→Glu255(氢键). 苯酚与FKBP5相互作用的结合能和结合面积分别为−3.91 kcal·mol−1和173 Å2.
如图2(c)所示,对苯二酚与FKBP5相互作用的关键氨基酸为Pro221、Glu227、Gly282、Gln286和Arg331. 对苯二酚与FKBP5的相互作用包括—OH→Gln286(氢键)、—OH→Pro221(氢键)、—OH→Glu227(氢键)、—OH→Gly282(氢键)和—OH←Arg331(氢键). 对苯二酚与FKBP5相互作用的结合能和结合面积分别为-3.81 kcal·mol−1和175 Å2.
如图2(d)所示,对苯醌与FKBP5相互作用的关键氨基酸为Lys204、Ala228、Met285、Arg331和Lys335. 对苯醌与FKBP5的相互作用包括C=O←Met285(氢键)、C=O←Arg331(氢键)、C=O←Ala228(氢键)、苯环-Ala228(范德华力相互作用)、C=O←Lys204(氢键)和C=O←Lys335(氢键). 对苯醌与FKBP5相互作用的结合能和结合面积分别为−3.83 kcal·mol−1和171 Å2.
如图2(e)所示,4-硝基邻苯二酚与FKBP5相互作用的关键氨基酸为Gly224、Asn238、Gly282、Met285和Gln286. 4-硝基邻苯二酚与FKBP5的相互作用包括—OH→Gly282(氢键)、—NO2 ←Met285(氢键)、—NO2←Asn238(氢键)、—OH→Gln286(氢键)和—OH→Gly224(氢键). 4-硝基邻苯二酚与FKBP5相互作用的结合能和结合面积分别为−3.69 kcal·mol−1和213 Å2.
如图2(f)所示,邻苯二酚与FKBP5相互作用的关键氨基酸为Gly224、Glu227、Ala228、Gly282、Tyr284和Gln286. 邻苯二酚与FKBP5的相互作用包括—OH→Gln286(氢键)、—OH→Gly224(氢键)、—OH→Gly282(氢键)、苯环-Ala228(范德华力相互作用)、—OH→Glu227(氢键)和苯环-Tyr284(π-π/范德华力相互作用). 邻苯二酚与FKBP5相互作用的结合能和结合面积分别为−3.81 kcal·mol−1和184 Å2.
-
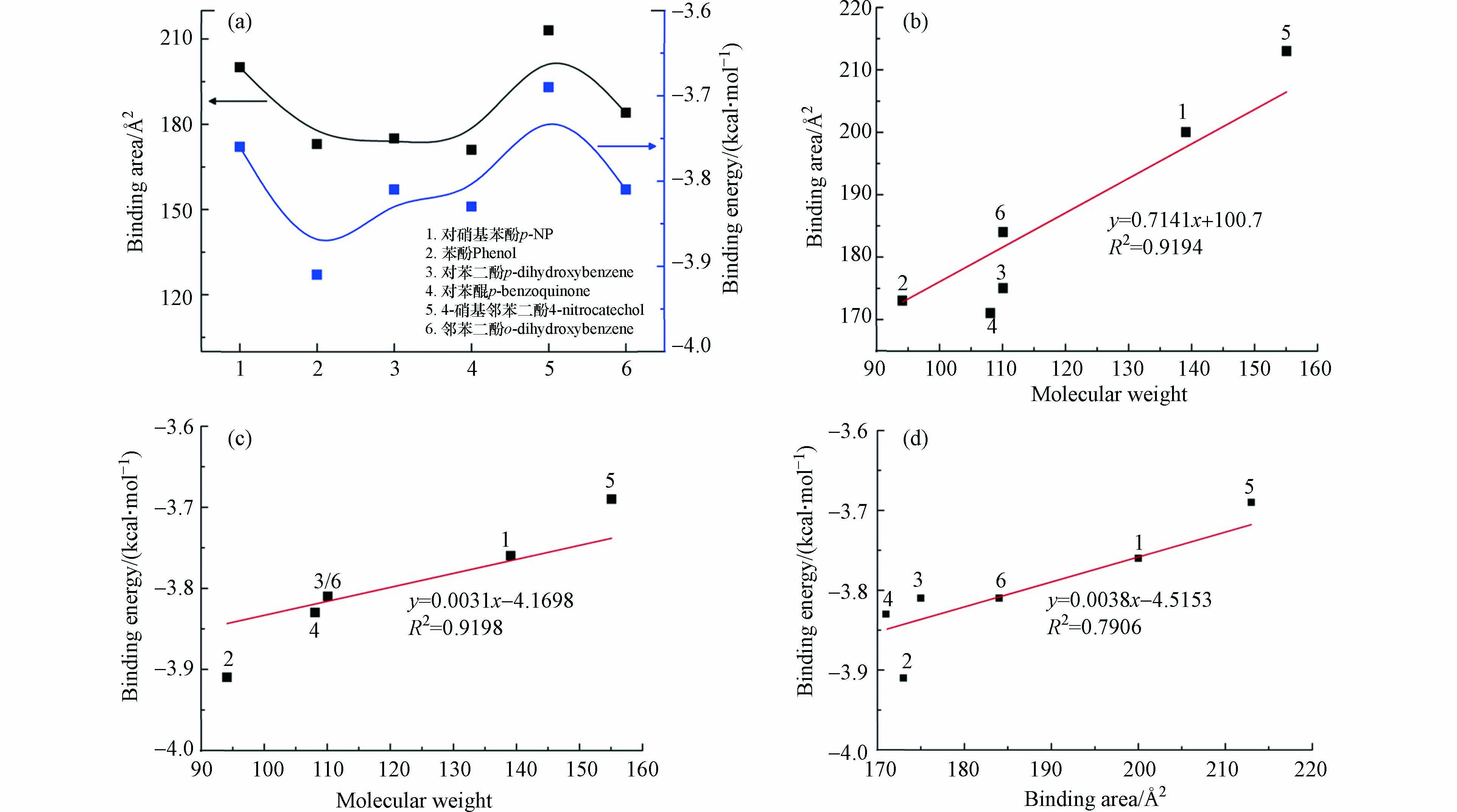
p-NP-DPs与FKBP5相互作用结果如图3(a)所示. p-NP-DPs与FKBP5相互作用的结合面积顺序为213 Å2(4-硝基邻苯二酚)> 200 Å2(p-NP)> 184 Å2(邻苯二酚)> 175 Å2(对苯二酚)> 173 Å2(苯酚)> 171 Å2(对苯醌),而p-NP-DPs与FKBP5相互作用的结合能顺序为−3.69 kcal·mol−1(4-硝基邻苯二酚)> −3.76 kcal·mol−1(p-NP)> −3.81 kcal·mol−1(对苯二酚/邻苯二酚)> −3.83 kcal·mol−1(对苯醌)> −3.91 kcal·mol−1(苯酚).
结合面积的数值越正意味着p-NP-DPs与FKBP5相互作用越强,而结合能的数值越负意味着p-NP-DPs与FKBP5相互作用越强,基于结合面积因素,p-NP-DPs与FKBP5相互作用强度大小顺序为4-硝基邻苯二酚 > p-NP > 邻苯二酚 > 对苯二酚 > 苯酚 > 对苯醌;基于结合能因素,p-NP-DPs与FKBP5相互作用强度大小顺序为苯酚 > 对苯醌 > 对苯二酚/邻苯二酚 > p-NP > 4-硝基邻苯二酚. 因此,结合能和结合面积对p-NP-DPs与FKBP5相互作用的影响是复杂的,p-NP-DPs与FKBP5相互作用的强度取决于结合能和结合面积复合作用的结果[28].
影响p-NP-DPs与FKBP5相互作用的关键因素可能是小分子的种类,而分子量是决定小分子性能和结构的关键因素,因此,研究p-NP-DPs分子量对p-NP-DPs与FKBP5相互作用的影响十分必要. p-NP-DPs分子量对p-NP-DPs与FKBP5相互作用结合面积的影响如图3(b)所示,结果表明p-NP-DPs分子量越大,p-NP-DPs与FKBP5相互作用结合面积越大,二者呈正相关关系,线性相关系数R2=0.9194,表明污染物分子量越大,污染物与受体蛋白相互作用越强,污染物环境风险越大. 而p-NP-DPs分子量对p-NP-DPs与FKBP5相互作用结合能的影响如图3(c)所示,p-NP-DPs分子量越大,p-NP-DPs与FKBP5相互作用结合能越大,二者呈正相关关系,线性相关系数R2=0.9198,表明污染物分子量越小,污染物与受体蛋白相互作用越强,污染物环境风险越大. p-NP-DPs与FKBP5相互作用结合面积和结合能的影响如图3(d)所示,结果表明结合面积和结合能呈正相关关系,线性相关系数R2=0.7906,因此分子量对p-NP-DPs与FKBP5相互作用强度影响随分子量增加呈“先升后降”趋势,过大的分子量导致污染物空间结构变大,受体蛋白空间位阻抑制p-NP-DPs与FKBP5相互结合,而过小的分子量则因为官能团和药效团减少抑制了p-NP-DPs与FKBP5相互结合[29 − 30].
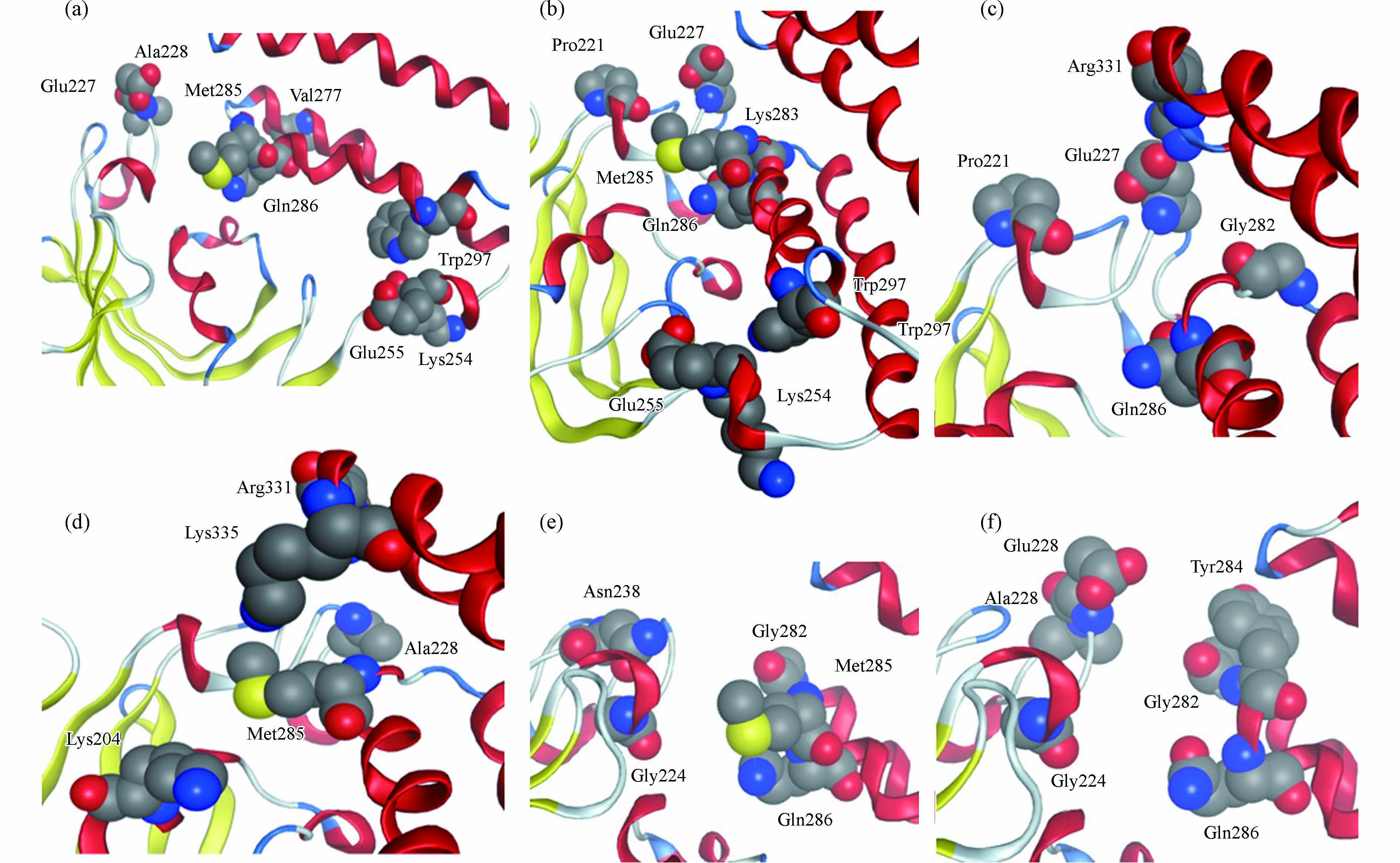
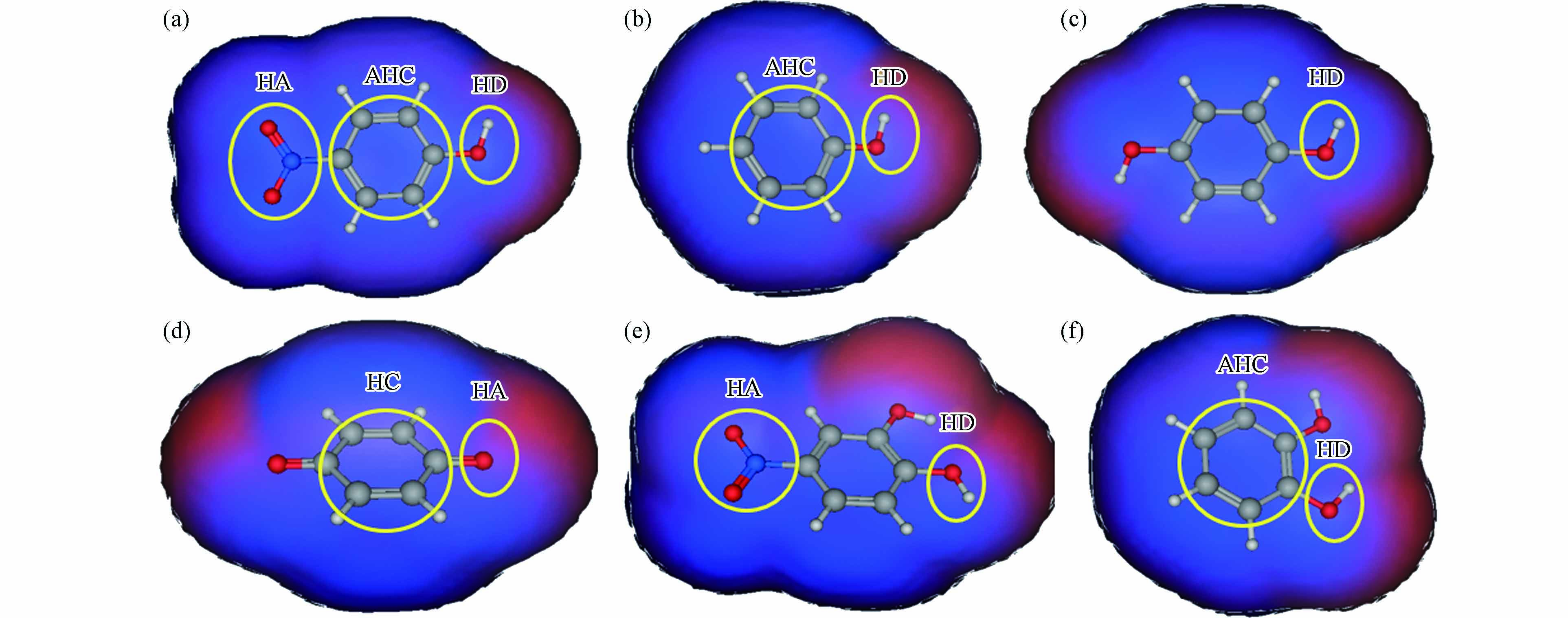
p-NP-DPs与FKBP5相互作用的结合区域如图4所示,p-NP-DPs药效团如图5所示. p-NP与FKBP5相互作用结合区域由Glu227、Ala228、Lys254、Glu255、Gly282、Met285、Gln286和Trp297组成,结合面积200 Å2(图4a),p-NP的药效团为氢键供体(—OH)、氢键受体(—NO2)和芳香中心-疏水中心(苯环及其C原子),如图5a所示. 苯酚与FKBP5相互作用结合区域由Pro221、Glu227、Lys254、Glu255、Lys283、Met285、Gln286和Trp297组成,结合面积173 Å2(图4b),苯酚的药效团为氢键供体(—OH)、芳香中心(苯环)和芳香中心-疏水中心(苯环上C原子),如图5b所示. 对苯二酚与FKBP5相互作用结合区域由Pro221、Glu227、Gly282、Gln286和Arg331组成,结合面积175 Å2(图4c),对苯二酚的药效团为氢键供体(—OH),如图5c所示. 对苯醌与FKBP5相互作用结合区域由Lys204、Ala228、Met285、Arg331和Lys335组成,结合面积171 Å2(图4d),对苯醌的药效团为氢键受体(C=O)和疏水中心(苯环),如图5d所示. 4-硝基邻苯二酚与FKBP5相互作用结合区域由Gly224、Asn238、Gly282、Met285和Gln286组成,结合面积213 Å2(图4e),4-硝基邻苯二酚的药效团为氢键供体(—OH)和氢键受体(—NO2),如图5e所示. 邻苯二酚与FKBP5相互作用结合区域由Gly224、Glu227、Ala228、Gly282、Tyr284和Gln286组成,结合面积184 Å2(图4f),邻苯二酚的药效团为氢键供体(—OH)和芳香中心-疏水中心(苯环),如图5f所示.
p-NP-DPs与FKBP5相互作用的氨基酸残基分布如图6所示,结果表明氨基酸残基分布区间为Lys204(对苯醌和FKBP5)到Lys335(对苯醌和FKBP5),其中高频氨基酸残基主要由两个区域组成,包括氨基酸残基序号221—228(Pro221、Gly224、Glu227、Ala228)和氨基酸残基序号282—286(Gly282、Lys283、Tyr284、Met285、Gln286).
-
p-NP-DPs分子结构的不同导致其理化性质(分子量、熔点、沸点、闪点、密度、lgSw、lgP、拓扑极表面积、氢键供体数量和氢键受体数量,表1)差异显著[31 − 32]. 为了明确p-NP-DPs与FKBP5相互作用强度与p-NP-DPs物理化学性质之间的关系,分别采用SPSS软件中的Spearman相关性和Pearson相关性分析了分子量、熔点、沸点、闪点、密度、lgSw、lgP、拓扑极表面积、氢键供体数量和氢键受体数量分别与结合面积和结合能绝对值之间的相关性. 表2为利用Spearman相关性分析理化性质对p-NP-DPs与FKBP5相互作用的影响,结果表明结合能绝对值与分子量和拓扑极表面积在0.01水平的相关系数均达到-1,与闪点、密度和氢键受体数量在0.05水平的相关系数达到-0.9747、-0.9747和-0.9549,表明结合能绝对值与分子量、拓扑极表面积、闪点、密度和氢键受体数量呈显著的负相关关系. 结合能绝对值与熔点、沸点、lgSw、lgP和氢键供体数量的相关系数分别为-0.6377、-0.7182、0.5000、-0.5798和-0.5323,且P>0.05,结合能绝对值与熔点、沸点、lgSw、lgP和氢键供体数量无相关性. 而结合面积仅与分子量和拓扑极表面积在0.05水平的相关系数均达到0.9276,表明结合面积仅与分子量和拓扑极表面积呈显著的正相关关系.
表3为利用Pearson相关性分析理化性质对p-NP-DPs与FKBP5相互作用的影响,结果表明结合能绝对值与分子量在0.01水平的相关系数达到−0.9591,与密度、拓扑极表面积和氢键受体数量在0.001水平的相关系数分别达到−0.9928、−0.9743和−0.9860,表明结合能绝对值与分子量、密度、拓扑极表面积和氢键受体数量呈显著的负相关关系. 结合面积与分子量、拓扑极表面积和氢键受体数量在0.01水平的相关系数达到0.9589、0.9597和0.9230,表明结合面积与分子量、拓扑极表面积和氢键受体数量呈显著的正相关关系. 结果表明,p-NP-DPs分子结构及理化性质对p-NP-DPs与FKBP5相互作用强度的影响主要取决于分子量和拓扑极表面积(100%)> 氢键受体数量(75%)> 密度(50%)> 闪点(25%).
-
(1)p-NP-DPs与FKBP5相互作用结合面积顺序为213 Å2(4-硝基邻苯二酚)> 200 Å2(p-NP)> 184 Å2(邻苯二酚)> 175 Å2(对苯二酚)> 173 Å2(苯酚)> 171 Å2(对苯醌),结合能顺序为−3.69 kcal·mol−1(4-硝基邻苯二酚)> −3.76 kcal·mol−1(p-NP)> −3.81 kcal·mol−1(对苯二酚/邻苯二酚)> −3.83 kcal·mol−1(对苯醌)> −3.91 kcal·mol−1(苯酚).
(2)p-NP-DPs与FKBP5相互作用高频氨基酸残基为Pro221、Gly224、Glu227、Ala228(221-228)和Gly282、Lys283、Tyr284、Met285、Gln286(282-286),p-NP-DPs的药效团主要包括氢键供体、氢键受体、芳香中心和疏水中心.
(3)p-NP-DPs理化性质对p-NP-DPs与FKBP5相互作用强度的影响主要取决于分子量和拓扑极表面积(100%)> 氢键受体数量(75%)> 密度(50%)> 闪点(25%).
对硝基苯酚降解产物与FK506结合蛋白相互作用分子机制
Molecular mechanism on interaction between p-nitrophenol degradation products and FK506 binding protein
-
摘要: 对硝基苯酚(p-nitrophenol,p-NP)作为典型内分泌干扰物,其环境污染与人体健康问题一直是环境领域的研究热点. 环境中对硝基苯酚降解产物(p-NP degradation products,p-NP-DPs)与FK506结合蛋白(FK506 binding protein,FKBP5)结合形成p-NP-DPs-FKBP5加合物是p-NP-DPs产生雌激素和抗雄激素活性的主要机制. 然而,p-NP-DPs与FKBP5相互作用的分子水平机制尚未引起足够的重视. 本文采用分子对接技术模拟计算p-NP-DPs与FKBP5相互作用的结合能和结合面积. 结果表明,p-NP-DPs与FKBP5相互作用结合面积顺序为213 Å2(4-硝基邻苯二酚)>200 Å2(p-NP)>184 Å2(邻苯二酚)>175 Å2(对苯二酚)>173 Å2(苯酚)>171 Å2(对苯醌),结合能顺序为−3.69 kcal·mol−1(4-硝基邻苯二酚)> −3.76 kcal·mol−1(p-NP)> −3.81 kcal·mol−1(对苯二酚/邻苯二酚)>−3.83 kcal·mol−1(对苯醌)> −3.91 kcal·mol−1(苯酚),高频氨基酸残基为Pro221、Gly224、Glu227、Ala228、Gly282、Lys283、Tyr284、Met285和Gln286,p-NP-DPs药效团主要包括氢键供体(—OH)、氢键受体(—NO2和C=O)、芳香中心(苯环)和疏水中心(C原子). p-NP-DPs理化性质对p-NP-DPs与FKBP5相互作用强度的影响主要取决于分子量和拓扑极表面积(100%)、氢键受体数量(75%)、密度(50%)和闪点(25%). 本研究对认识水环境中p-NP-DPs分子水平健康效应和环境风险具有重要科学意义.Abstract: P-nitrophenol (p-NP), as typical endocrine disruption chemicals (EDCs), their environmental pollution and human health issues have always been hotspots in the environmental field. The p-NP-DPs-FKBP5 adducts, formed by p-NP degradation products (p-NP-DPs) and FK506 binding protein (FKBP5), is the main estrogenic and anti-androgenic mechanism. However, this molecular-level mechanism between p-NP-DPs and FKBP5 has not attracted enough attention. In this study, the molecular docking technology was used to simulate the binding energy and binging area between p-NP-DPs and FKBP5 interaction. The results indicated that the order of binging area between p-NP-DPs and FKBP5 interaction was 213 Å2 (4-nitrocatechol) > 200 Å2 (p-NP) > 184 Å2 (o-dihydroxybenzene) > 175 Å2 (p-dihydroxybenzene) > 173 Å2 (phenol) > 171 Å2 (p-benzoquinone), but the order of binding energy between p-NP-DPs and FKBP5 interaction was −3.69 kcal·mol−1 (4-nitrocatechol) > −3.76 kcal·mol−1 (p-NP) > −3.81 kcal·mol−1 (p-/o-dihydroxybenzene) > −3.83 kcal·mol−1 (p-benzoquinone) > −3.91 kcal·mol−1 (phenol). The high frequency amino acid residues were included Pro221, Gly224, Glu227, Ala228, Gly282, Lys283, Tyr284, Met285 and Gln286. The pharmacophores of p-NP-DPs were included hydrogen-bond donor (—OH), hydrogen-bond acceptor (—NO2 and C=O), aromatic center (benzene ring) and hydrophobic center (C atom). The physicochemical properties of p-NP-DPs were dependent on molecular weight and topological polar surface area (100%), hydrogen bond donor count (75%), density (50%) and flash point (25%). The research results probably enhance people’s knowledge and understanding of molecular-level health effects and environmental risks of p-NP-DPs in the water environmental system in the future.
-
Key words:
- p-NP /
- FKBP5 /
- molecular docking /
- pharmacophore /
- structure-activity relationship
-
黑臭水体是城市化进程中水生系统面临的一个重要的环境问题. 2015年国务院发布的《水污染防治行动计划》(简称“水十条”)中明确要求,采取控源截污、垃圾清理、清淤疏浚、生态修复等措施,加大黑臭水体治理力度[1]. 其中生态修复主要利用微生物、水生动植物等生物的生命活动,对水中污染物进行转移、转化及降解,恢复水体在一定污染负荷下的自净化能力[2]. 在水生态修复工作中,恢复水生植物尤其是恢复沉水植物被广泛认为是水体治理的有效途径[3]. 目前用于恢复河湖生态的沉水植物种植常通过排干上覆水、清淤后将沉水植物种植于水体土壤中,再蓄水使沉水植物生长达到净化水质、防治污染的目的. 但该种植方式耗时长、成本高,且河湖清淤后的底质较硬,植物生长必需的营养物质也随清淤过程被移除,导致沉水植物成活率较低. 另外,部分城市硬化沟渠因其“三面光”的特点,在发生黑臭后的生态修复主要依靠生态浮岛种植浮水植物,而沉水植物需要沉没于水中完成生活史,其通过光合作用产生的氧气都释放到水中,可以显著增加水体中的溶解氧含量[4]. 因此,开发新型种植技术提高沉水植物的河湖生态修复效果具有重要意义.
针对沉水植物种植困难的问题,目前有基于砾石和滤料构成的生物循环床[5]、网床或网箱[6],以及基于苯乙烯和树脂构成的生态浮岛[7]. 而这些基质都存在经济和环境效益问题,例如维护成本高、微塑料释放等. 污泥陶粒是污泥稳定化、轻量化及无害化处置的产物,具有比表面积大和多孔结构等特性. 据统计,至2021年全国已建成
2827 座城市污水处理厂,每年产生的污泥量约3000 —6000 万t[8]. 我国污水处理产业因处理能力、技术和投入仍存在不足,行业内“重水轻泥”的现状依然没有得到有效改善,导致大量污泥未能得到有效处置[9]. 目前研究报道的污泥陶粒基于其多孔结构的特性已被广泛用作建筑材料[10]、人工湿地基质[11 − 13]、滤池填料[14 − 16]和各种水处理设备的滤材[17]. 已报道的污泥陶粒中,吸水率大多在50% 以下,导致其应用于人工湿地时需补充土壤或砂砾才能满足湿地植物的种植[12]. 而亲水性污泥陶粒保水保墒更利于植物的定植和生根,在提供固着基质的同时,还能避免植物烧根[17]. 同时亲水性污泥陶粒的吸附性能使得其可为植物根系提供营养物质,更利于植物的生长发育. 通过污泥陶粒构建的人工湿地用于污水处理后,能值产出和可持续性指数相较其他污泥处置方式更高[11]. 因此,开发利用亲水性污泥陶粒种植水生植物不仅能解决沉水植物种植困难的问题,还能进一步实现污泥资源化利用,具有可观的经济效益,符合可持续性发展的需求.基于此,本研究提出亲水性污泥陶粒可作为水生植物的栽培基质(“新型土壤”),提高水生植物存活率. 开发一套运用亲水性污泥陶粒种植沉水植物构建“水下森林”生态系统的新策略,以恢复淡水生态系统的结构和功能并改善水质. 该策略的实施将为城市淡水生态系统实现固碳增汇,为“双碳”目标的实现提供新思路.
1. 实验部分(Experimental section)
1.1 亲水性污泥陶粒的准备和表征
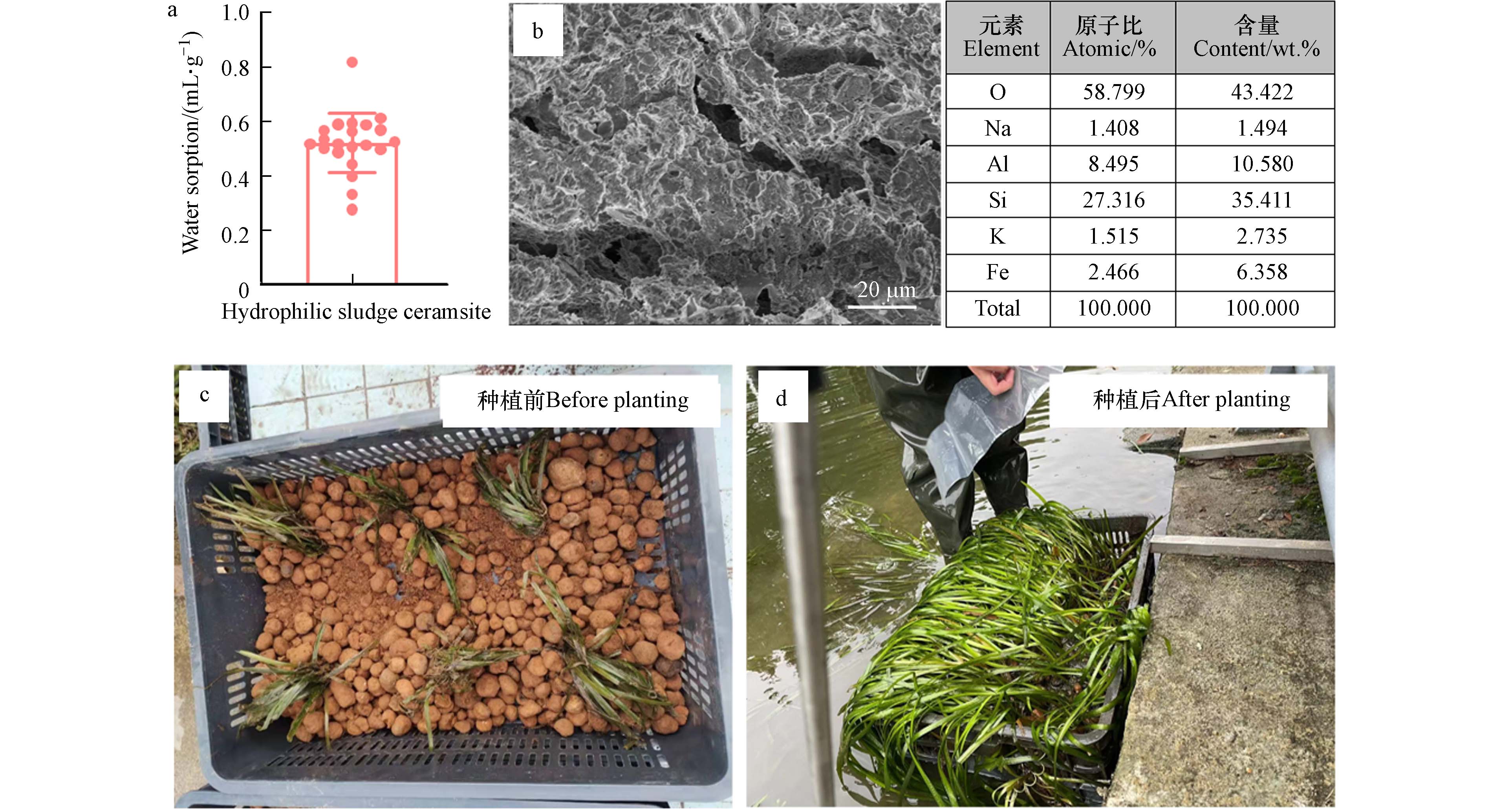
亲水性污泥陶粒购自武汉铭创新海生态科技有限公司,公司基于专利“一种高掺量污泥蓄水材料的制备工艺,CN
114920541 A”进行制备,具体制备流程为将污泥、秸秆和添加剂按比例混合均匀后经过造粒及造粒后在窑炉中1000 ℃烧结20 min制备而成. 亲水性污泥陶粒的形貌结构用Hitachi SU8000扫描电子显微镜(SEM)和能谱仪(EDS)进行表征. 吸水率由公式(1)得出:吸水率=V0−Vm (1) 其中V0为溶液初始体积,V为亲水性污泥陶粒吸水后溶液的体积,m为亲水性污泥陶粒的质量.
1.2 沉水植物的种植及监测分析
“水下森林”的构建由灰色聚丙烯材质的网篮作为固定单元,装入一定质量的亲水性污泥陶粒,并将苦草(Vallisneria natans (Lour.) H. Hara)埋入亲水性污泥陶粒中5 cm,将种植模块整体沉入水中.
每个种植模块种植固定质量的沉水植物,选取3个种植模块进行标记并记录沉水植物的具体质量和叶长. 经过生长两年后,取出标记模块,收集模块中所有沉水植物后,返回实验室对收集到的沉水植物进行清洗,去除根系附着的土壤等,自然沥干1 h后进行称重和叶长记录.
1.3 示范区域及样品采集
江汉大学校河—清源河呈“C”型,两端分别联通于三角湖,有闸口可控制三角湖水的输入与排放,全长约1 km,水深约1—1.3 m,河道经水泥硬化处理. 作为校园雨水和生活污水的汇集地,清源河存在许多外源氮、磷输入,并且由于清源河的水泥硬化人工河道,导致水体自净能力不足,进而水质逐渐恶化. 自2021年7月分别于3#和4#号点位种植了苦草构建淡水生态系统,并设置5个取样点(1#、2#、3#、4#、5#)监测水质变化. 取样点分布如图1所示.
为探究种植沉水植物后微生物群落的变化,分别在无草水域、少草水域和多草水域采亲水性污泥陶粒样本,取亲水性污泥陶粒内(距中心点0.5 cm半径以内材料)外(距中心点0.5 cm半径以外的材料)样本. 每个样本准备3个重复样品,共18个样品进行16S rRNA测序分析.
1.4 水体样品检测
本研究测定了清源河5个监测点的 6项水质指标. 其中,溶解氧(DO)采用化学探头法(HJ 506-2016)(便携式多参数水质分析仪,YSIproQuatro)测定;高锰酸盐指数(CODMn)采用酸性法(GB/T
11892 -1989)(连续数字滴定仪,Titrette)测定;氨氮(NH4+-N)采用纳氏试剂分光光度法(HJ 535-2009)(紫外可见分光光度计,UV-7504 )测定;硝酸盐氮(NO3−-N)采用离子色谱法(HJ 84-2016)(离子色谱,AQ-1100 )测定;总氮(TN)采用碱性过硫酸钾消解紫外分光光度法(HJ 636-2012)(紫外可见分光光度计,UV-1800PC);总磷(TP)采用钼酸铵分光光度法(GB/T11893 -1989)(紫外可见分光光度计,UV-7504 )测定.1.5 微生物群落分析
使用TGuide S96 Magnetic Soil /Stool DNA Kit (天根生化科技(北京)有限公司),按照说明书,从18个样品中提取全基因组DNA. 用引物338F: 5’- ACTCCTACGGGAGGCAGCA-3’和806R: 5’- GGACTACHVGGGTWTCTAAT-3’扩增细菌16S rRNA高变区V3—V4. PCR产物在琼脂糖凝胶上检测,并通过Omega DNA纯化试剂盒(Omega Inc., Norcross, GA, USA)纯化. 收集纯化的PCR产物,在Illumina Novaseq
6000 平台上进行2 × 250 bp的配对. 使用USEARCH将相似阈值大于97%的符合条件的序列分配到一个操作分类单元(OTU). 数据结果上传到国家生物信息中心https://ngdc.cncb.ac.cn/bioproject/,项目编号PRJCA021118,GSA编号CRA014420. 基于QIIME2中的朴素贝斯分类器,使用SILVA数据库对OTUs/ASV进行分类标注,置信阈值为70%. 利用QIIME2软件对各样本物种多样性复杂性进行Alpha鉴定. 采用主坐标分析方法计算Beta多样性,评价样品的物种复杂性. 采用单因素方差分析比较细菌丰度和多样性.2. 结果与讨论(Results and discussion)
2.1 亲水性污泥陶粒结构表征
高温烧制的亲水性污泥陶粒具备了吸水性、多孔结构、比表面积大等特性. 其中,亲水性污泥陶粒的球体结构组成的间隙利于沉水植物的生根和固着,能有效提高沉水植物在恶劣环境的存活率,同时避免传统种植方式中营养元素过剩导致的植物烧根现象[17]. 本研究采用的亲水性污泥陶粒的吸水率可达0.522 mL·g−1(图2 a),这为微生物定殖和物质交换提供了良好的基础. 亲水性污泥陶粒的扫描电镜图像显示其具有丰富的孔径,且元素组成与土壤极为相似,主要含有氧、硅、铝等元素(图2 b). 因此,亲水性污泥陶粒的亲水性和多孔结构可以实现吸附营养物质、过滤悬浮物、为微生物和小型浮游动物提供栖息地,固着沉水植物根系并提供营养物质等功能. 本研究以网篮作为固定单元,亲水性污泥陶粒作为种植基质种植沉水植物,种植方式见图2 c. 经过两年时间的生长,沉水植物不仅存活率高,且长势良好(图2 d). 虽然高温烧制去除了污泥中原有的大部分氮、磷、有机质等植物生长必需物质,但其特有的亲水性和多孔结构使亲水性污泥陶粒可以从水体中有效吸附氮、磷、有机质、微量元素等维持水生植物的正常生长[17]. 水生植物的光合作用可以为微生物和浮游生物提供氧气,微生物和浮游生物呼吸作用产生的二氧化碳和代谢产生的有机物可以被水生植物吸收利用,实现正向的生态循环[18].
2.2 沉水植物生长
本文将亲水性污泥陶粒作为水生植物的栽培基质(“新型土壤”),用于种植水生植物,恢复淡水生态系统的结构和功能. 不同于传统的将沉水植物直接种植在河湖底泥的方式,利用陶粒作为“新型土壤”种植沉水植物仅需在固定装置中装满陶粒,将沉水植物根部栽种至陶粒间隙后即可通过不同的方式将种植模块放置于河湖渠底部,实现沉水植物的高效种植(图2 c). 本研究中,苦草种植前后的分蘖数及生物量显著增多. 每框植物的平均重量从种植前的241.20 g显著增长至
1566.13 g (图3 a),植物叶长从40 cm增长至120 cm (图3 b). 且经过两年的定植,植物能分蘖出2—3株新植株,新分蘖的植株最长能到60 cm (图3 c和d). 上述结果表明亲水性污泥陶粒种植模块利于沉水植物的定植. 亲水性污泥陶粒种植沉水植物的方法相较于传统种植更高效、经济. 这种方式无需排干河湖,简化了种植过程,节省了时间和成本. 其模块化设计使得维护更为便捷,仅需替换长势不良的模块,而无需大规模更换植物或排干水体.2.3 水质变化分析
自2021年7月于3#和4#点位用亲水性污泥陶粒种植模块种植了苦草,并设置1#、2#和5#点位作为对照(图4 a),以此探究该种植方式构建新淡水生态系统的潜力. 至2023年5月沉水植物生长状态良好(图4 b). 自2022年2月起对3#号种植点和2#号对照点进行了生态系统拍摄(图4 d-g). 由图4 b和c可看出,相较于种植沉水植物之前,亲水性污泥陶粒种植的沉水植物生长正常,水体透明度高,并基本构建了清源河“水下森林”新淡水生态系统. 从2022年2月至2023年12月的一年多时间里,种植区的沉水植物生长正常,水体透明度及水质状况呈现持续向好的趋势,生态系统保持较好(图4 e,g,3#号点),未种植沉水植物的水域始终浑浊(图4 d,f,2#号点).
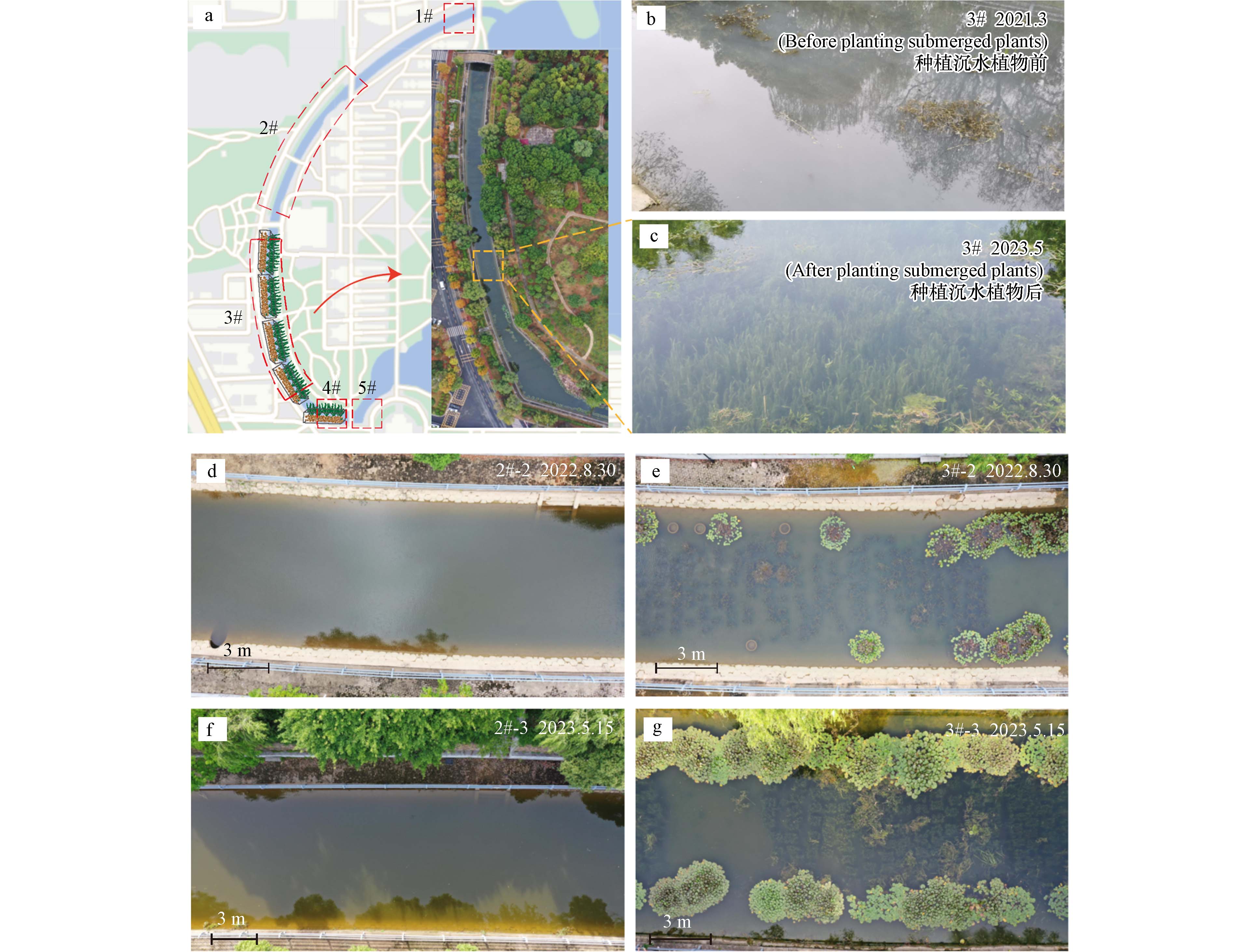 图 4 亲水性污泥陶粒种植沉水植物构建“水下森林”淡水生态系统种植点和对照点示意图(a),种植前(b)后(c)水质情况,2022—2023年对照点(d,f)和种植点(e,g)的水质变化Figure 4. Construction of ‘underwater forest’ freshwater ecosystem by planting submerged plants with hydrophilic sludge ceramsite schematic diagram of planting sites and control sites (a), water quality before (b) and after (c) planting, water quality changes at control sites (d, f) and planting sites (e, g) from 2022 to 2023
图 4 亲水性污泥陶粒种植沉水植物构建“水下森林”淡水生态系统种植点和对照点示意图(a),种植前(b)后(c)水质情况,2022—2023年对照点(d,f)和种植点(e,g)的水质变化Figure 4. Construction of ‘underwater forest’ freshwater ecosystem by planting submerged plants with hydrophilic sludge ceramsite schematic diagram of planting sites and control sites (a), water quality before (b) and after (c) planting, water quality changes at control sites (d, f) and planting sites (e, g) from 2022 to 2023为评估亲水性污泥陶粒种植沉水植物构建“水下森林”新淡水生态系统用于改善水质的实际效果,分别于秋季(2022年11月)、冬季(2023年2月)和春季(2023年5月)测定了种植点和对照点水质相关参数. 溶解氧(DO)、总磷(TP)、总氮(TN)、氨氮(NH4+-N)和硝酸盐氮(NO3−-N)的结果表明,沉水植物种植点位(3#和4#点)的DO含量为明显高于对照点,且呈现了随种植时间递增的趋势,至2023年春季水体DO含量可达11.47 mg·L−1,有益于水生生物的生存及污染物的降解;而TP、TN、NH4+-N、NO3−-N浓度低于其它点位,且呈现了明显的下降趋势(表1). 污染物浓度上,基于国家地表水环境质量标准(GB
3838 -2002)[19],运用亲水性污泥陶粒种植了沉水植物的点位(3#和4#点)较对照点相比,总体水质已从劣V类提升至近III类(表1). 其中,水体TN从劣V类(2022年11月2#点,> 8.0 mg·L−1)水质恢复至近III类(2023年5月4#点,< 1.0 mg·L−1);NH4+-N从IV类(2022年11月2#点,> 5.0 mg·L−1)水质恢复至近I类(2023年5月3#和4#点,< 0.15 mg·L−1);TP从劣V类(2022年11月1#点,> 0.5 mg·L−1)水质恢复至III类/近II类(2023年5月3#和4#点,< 0.05 mg·L−1);高锰酸钾指数(CODMn)提升至II类(2023年5月4#点,< 2.5 mg·L−1)水质;DO提升至I类水质(2023年5月3#和4#点,> 11 mg·L−1)(表1). 以上结果表明,亲水性污泥陶粒种植沉水植物构建“水下森林”新淡水生态系统能有效改善河湖水质,实现了污泥资源化产物的再利用和黑臭水体的生态修复.表 1 清源河采样点主要污染指标与地表水环境质量的比较Table 1. Comparison of main pollution index and surface water environmental quality in Qingyuan River sampling sites点位Sites 时间Time DO/(mg·L−1) TP/(mg·L−1) TN/(mg·L−1) NH4+-N/(mg·L−1) NO3−-N/(mg·L−1) CODMn/(mg·L−1) 1#(对照点)Control site 2022.11 8.10 0.39 6.92 2.51 1.58 5.60 2023.02 8.37 0.08 1.93 0.71 0.84 3.28 2023.05 5.88 0.14 1.80 1.38 0.53 5.30 2#(对照点)Control site 2022.11 5.37 0.56 8.87 5.30 1.34 5.80 2023.02 10.69 0.05 1.17 0.39 0.55 4.12 2023.05 7.49 0.12 0.97 0.36 0.33 3.50 3#(种植点)Planting site 2022.11 6.33 0.16 2.91 1.02 1.12 4.30 2023.02 10.06 0.08 1.28 0.43 0.44 2.96 2023.05 11.36 0.03 1.09 0.13 0.76 3.50 4#(种植点)Planting site 2022.11 5.59 0.12 1.33 0.43 0.83 3.30 2023.02 10.36 0.06 1.19 0.22 0.13 3.20 2023.05 11.47 0.03 0.69 0.12 0.46 2.40 5#(对照点)Control site 2022.11 2.74 0.50 6.79 4.45 0.22 8.00 2023.02 6.02 0.43 6.94 4.92 1.13 4.08 2023.05 6.93 0.23 1.80 1.43 0.53 4.10 I类[19] — ≥ 7.5 ≤ 0.01 ≤ 0.2 ≤ 0.15 — ≤ 2.0 II类[19] — ≥ 6 ≤ 0.025 ≤ 0.5 ≤ 0.5 — ≤ 4.0 III类[19] — ≥ 5 ≤ 0.05 ≤ 1.0 ≤ 1.0 — ≤ 6.0 IV类[19] — ≥ 3 ≤ 0.1 ≤ 1.5 ≤ 1.5 — ≤ 10 V类[19] — ≥ 2 ≤ 0.2 ≤ 2.0 ≤ 2.0 — ≤ 15 2.4 微生物群落结构及生物多样性
沉水植物可调节水体中溶解氧的动态改变,水体中溶解氧的升高则是影响生态重构和生物多样性的关键因素[20]. 考虑到黑臭水体的形成与厌氧菌大量繁殖直接相关[21],本文分析了亲水性污泥陶粒内外定殖的微生物群落结构及生物多样性. 分别在无草水域、少草水域和多草水域采污泥陶粒样本,经过16S rRNA测序得到
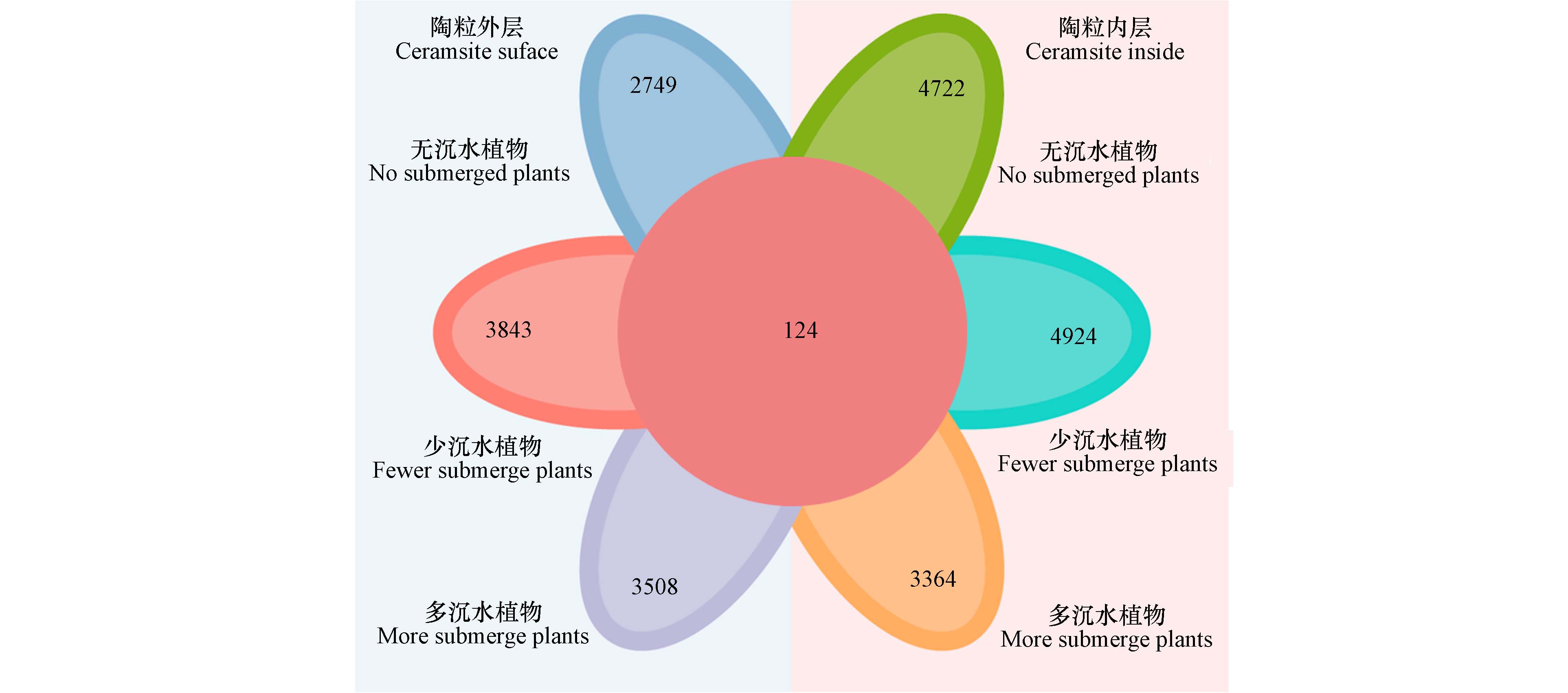
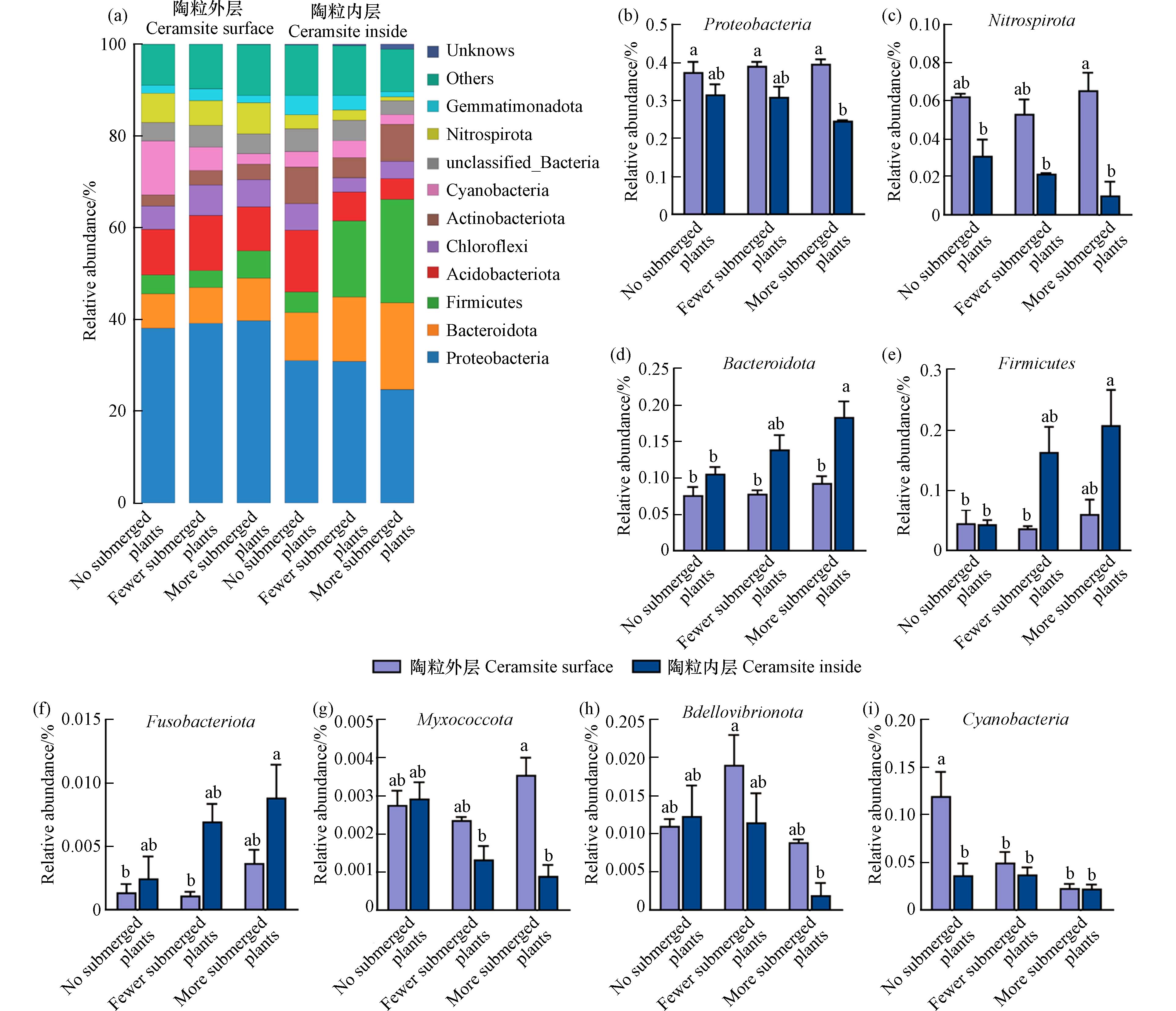
26731 个分类单元(operational taxonomic units,OTU). 其中,无草水域的污泥陶粒外层有2749 个OTU,少于其内层(4722 个OTU). 少草水域的污泥陶粒外层有3843 个OTU,少于其内层(4924 个OTU). 多草水域的污泥陶粒外层有3508 个OTU,略多于其内层(3364 个OTU)(图5). 结果表明无论是否存在沉水植物,污泥陶粒内层定殖的微生物种类均多于外层,这表明亲水性污泥陶粒的球体结构适合微生物的栖息和繁殖. 在物种分布柱状图中,研究结果得到了污泥陶粒的优势细菌门,分别为变形菌门(Proteobacteria),拟杆菌门(Bacteroidota),厚壁菌门(Firmicutes),酸杆菌门(Acidobacteriota),绿弯菌门(Chloroflexi),放线菌门(Actinobacteriota),蓝藻门(Cyanobacteria),螺旋硝化菌门(Nitrospirota)和芽单胞菌门(Gemmatimonadota)(图6 a). 其中,好氧菌属的变形菌门(Proteobacteria)和硝化螺旋菌门(Nitrospirota)在污泥陶粒外部相对丰度较高,且在多草水域的污泥陶粒内外差异显著(图6 b和c). 专性厌氧的拟杆菌门(Bacteroidota)、梭杆菌门(Fusobacteriota)和多为厌氧菌的厚壁菌门(Firmicutes)在污泥陶粒内部的相对丰度较多,且随着沉水植物的增多,污泥陶粒内外差异逐渐显著(图6 d,e和f). 捕食性微生物作为微生物群落中的捕食者,在维持微生物群落的多样性及生态功能中发挥重要作用[22-23]. 由图6 g、h可知,在沉水植物种植水域的污泥陶粒中,黏细菌(Myxococcota)和蛭弧菌(Bdellovibrionota)等捕食性微生物更倾向于分布在污泥陶粒的表层,水体中溶解氧的升高可直接影响污泥陶粒表层和内部微生物群落结构的组成和功能. 但沉水植物过多后,无论是污泥陶粒外层还是内层,少草水域的污泥陶粒OTU数均多于无草和多草水域的污泥陶粒,这说明在溶解氧和植物化感作用的共同影响下,微生物群可能存在定向演替[4, 24].除此之外,蓝藻(Cyanobacteria)作为水华的典型生物[25-26],在无草水域的陶粒外层定殖较多,而少草和多草水域污泥陶粒中无论内层还是外层相对丰度显著降低(图6 i),证实种植沉水植物对蓝藻具有明显的抑制作用,这可能是由于苦草等对蓝藻的化感作用[27].
2.5 “水下森林”生态修复作用过程
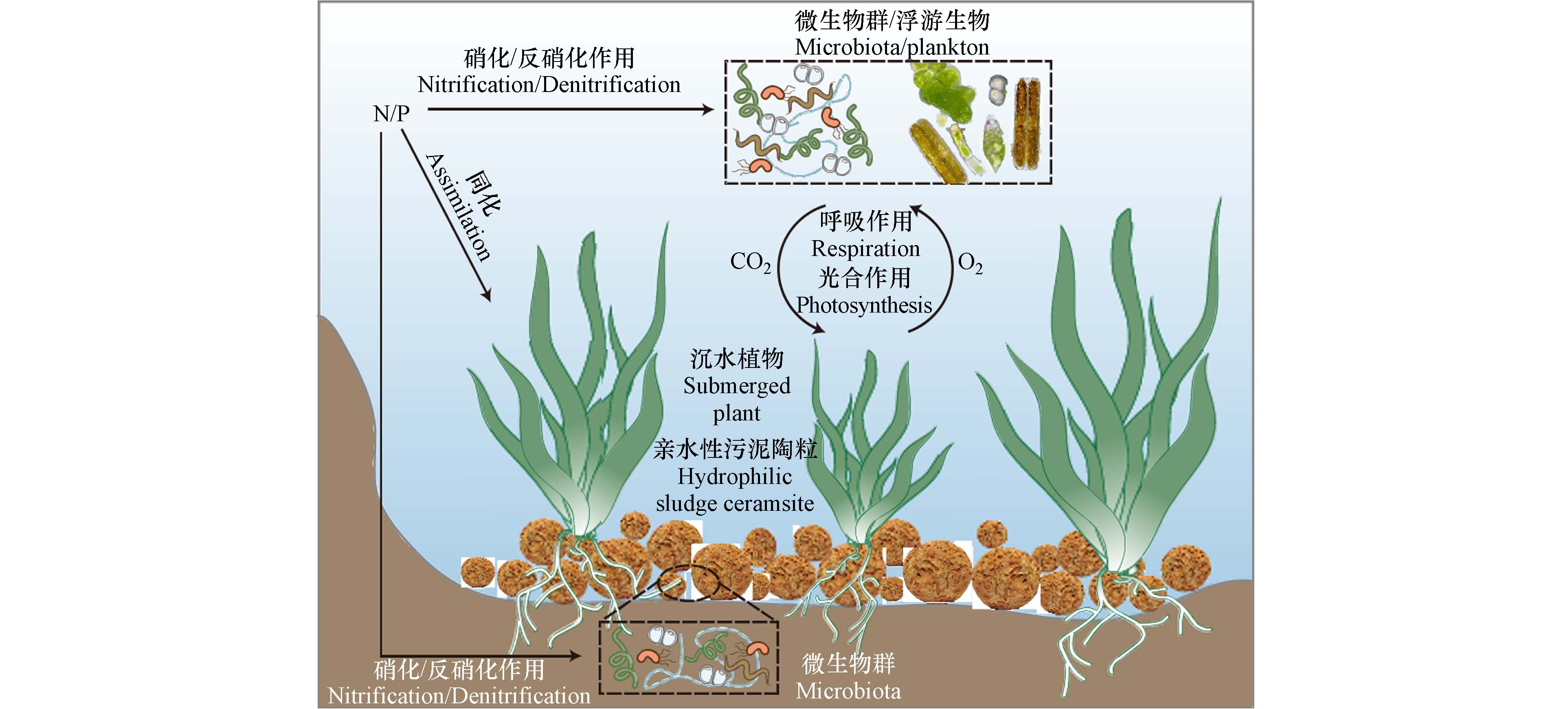
基于亲水性污泥陶粒开展城市黑臭水体修复及生态重构的应用示范取得了良好的示范效果,归纳总结了其作用机理如图7所示. 一方面,亲水性污泥陶粒为沉水植物提供了固着基质,且沉水植物可有效提升水体溶解氧浓度,有利于水生微型生物群落的重构和稳定;另一方面,亲水性污泥陶粒的引入为厌氧微生物和兼性厌氧微生物提供了适宜的微生境,陶粒内部捕食性微生物的分布趋势和蓝藻的生长抑制,均证实溶解氧的升高对水生微型生物的结构和功能具有关键性作用,示范水体中氮磷的去除及转化规律也进一步证实水体中微型生物的多样性是控制水质的核心指标. 亲水性污泥陶粒用于定植沉水植物的“水下森林”重构策略有利于恢复水体生物多样性,水体中溶解氧的提升,促进了轮虫、桡足类、枝角类等浮游动物的生存,并通过“下行效应”有效抑制水体富营养化—藻华—黑臭水体的演化过程,最终实现城市黑臭水体的原位生态修复.
从淡水生态系统角度出发,基于亲水性污泥陶粒的沉水植物种植模块可以实现沉水植物的成功种植和后期繁殖,并为浮游生物和微生物提供栖息地,从而提高水生态系统的生物多样性. 其中的沉水植物可以吸收水体中的营养盐和二氧化碳,促进各种元素循环,同时为定殖的微生物和浮游生物提供氧气. 而定殖的微生物形成的生物膜通过硝化反硝化作用进行氮的转化利用,浮游生物则通过营养级联效应制衡微生物,实现水体生物多样性的有效恢复,预防黑臭水体的发生和生态功能的丧失.
从元素循环角度出发,碳、氮、磷、硫等元素可以通过沉水植物及定殖的微生物进行转化,并通过食物链达到平衡. 在底泥/污泥的无害化处置过程中,烧制陶粒过程会消耗能源排放废气,属于“增”碳过程,而构建“水下森林”进行水生态修复为“减”碳过程,本研究开发的新策略可为国家达成“碳中和”目标提供了新思路和技术支撑.
3. 结论(Conclusion)
本文利用污泥烧结成的亲水性污泥陶粒种植沉水植物,重构了“水下森林”淡水生态系统,达到了改善水质和恢复生态的目的,实现了城市生态系统的污泥经烧制成陶粒最终又应用到淡水生态系统修复的无害化、资源化循环利用. 重建水生态系统之后的清源河水质从劣V类提升至近III类,各指标提升明显,水体透明度显著提高. 亲水性污泥陶粒定殖微生物的结果表明,亲水性污泥陶粒可以调控微生物的群落分布,并且沉水植物的化感作用会影响微生物群落的定向演替. 沉水植物光合作用产生的氧气能有效提升水体环境DO,抑制了因厌氧微生物过量繁殖导致的水体变黑变臭.
-
图 5 p-NP-DPs与FKBP5相互作用药效团:p-NP(a)、苯酚(b)、对苯二酚(c)、对苯醌(d)、4-硝基邻苯二酚(e)和邻苯二酚(f)氢键受体: hydrogen-bond acceptor(HA);氢键供体: hydrogen-bond donor(HD);芳香-疏水中心: aromatic-hydrophobic center(AHC);疏水中心: hydrophobic center(HC)
Figure 5. Pharmacophores between p-NP-DPs and FKBP5: p-NP (a), phenol (b), p-dihydroxybenzene (c), p-benzoquinone (d), 4-nitrocatechol (e) and o-dihydroxybenzene (f)
表 1 p-NP-DPs的理化性质
Table 1. Physicochemical property of p-NP-DPs
名称Sample CAS 熔点/°C Melting point 沸点/°C Boiling poin 闪点/°C Flash point 密度/(g·cm−3) Density lgSw p-NP 100—02—7 113—114 279 169 1.480—1.50 -0.74 苯酚Phenol 108—95—2 40.9 181.8 79 1.04—1.07 0 对苯二酚p-dihydroxybenzene 123—31—9 170—173 285—287 165 1.330—1.332 N/A 对苯醌p-benzoquinone 106—51—4 114—116 180 93 1.318—1.320 N/A 4-硝基邻苯二酚4-nitrocatechol 3316—09—4 174—176 N/A N/A N/A N/A 邻苯二酚o-dihydroxybenzene 120—80—9 105 245 127 1.344 0.62 名称Sample 相对分子质量Molecular weight lgP 拓扑极表面积/Å2Topological polar surface area 氢键供体数量Hydrogen bond donor count 氢键受体数量Hydrogen bond acceptor count p-NP 139.1100 1.91 66 1 3 苯酚Phenol 94.1130 1.46 20.2 1 1 对苯二酚p-dihydroxybenzene 110.1120 0.59 40.5 2 2 对苯醌p-benzoquinone 108.0960 0.2 34.1 0 2 4-硝基邻苯二酚4-nitrocatechol 155.1090 1.66 86.3 2 4 邻苯二酚o-dihydroxybenzene 110.1120 0.88 40.5 2 2 说明:N/A为数据未查到 表 2 p-NP-DPs与FKBP5相互作用和理化性质Spearman相关性分析
Table 2. Spearman correlation between physicochemical property and interaction of p-NP-DPs and FKBP5
理化性质Physicochemical property 结合能Absolute value of binding energy 结合面积Binding area 相关性系数Correlation coefficient P 相关性系数Correlation coefficient P 分子量Molecular weight −1** 0.0056 0.9276* 0.0167 熔点Melting point −0.6377 0.2000 0.3714 0.4972 沸点Boiling point −0.7182 0.1667 0.7000 0.2333 闪点Flash point −0.9747* 0.0333 0.8000 0.1333 密度Density −0.9747* 0.0333 0.9000 0.0833 lgSw 0.5000 1.0000 −0.5000 1.0000 lgP −0.5798 0.2389 0.7714 0.1028 拓扑极表面积Topological polar surface area −1** 0.0056 0.9276* 0.0167 氢键供体数量Hydrogen bond donor count −0.5323 0.3000 0.6172 0.2333 氢键受体数量Hydrogen bond acceptor count −0.9549* 0.0167 0.8197 0.0667 * P < 0.1,** P < 0.05. 表 3 p-NP-DPs与FKBP5相互作用和理化性质Pearson相关性分析
Table 3. Pearson correlation between physicochemical property and interaction of p-NP-DPs and FKBP5
理化性质Physicochemical property 结合能Absolute value of binding energy 结合面积Binding area 相关性系数Correlation coefficient P 相关性系数Correlation coefficient P 分子量Molecular weight −0.9591** 0.0025 0.9589** 0.0025 熔点Melting point −0.7880 0.0626 0.4672 0.3502 沸点Boiling point −0.7669 0.1303 0.6279 0.2567 闪点Flash point −0.8471 0.0701 0.6638 0.2218 密度Density −0.9928*** 0.0000 0.6887 0.1985 lgSw 0.3750 0.7553 −0.6296 0.5665 lgP −0.3641 0.4780 0.7306 0.0991 拓扑极表面积Topological polar surface area −0.9743*** 0.0000 0.9597** 0.0024 氢键供体数量Hydrogen bond donor count −0.4120 0.4170 0.4180 0.4095 氢键受体数量Hydrogen bond acceptor count −0.9860*** 0.0000 0.9230** 0.0087 ** P < 0.05,*** P < 0.01. -
[1] 李易, 陆锐, 沈锦优, 等. 废水中硝基芳香化合物检测方法研究进展[J]. 环境化学, 2016, 35(7): 1474-1485. doi: 10.7524/j.issn.0254-6108.2016.07.2015113001 LI Y, LU R, SHEN J Y, et al. Detection methods for nitroaromatic compounds in wastewater[J]. Environmental Chemistry, 2016, 35(7): 1474-1485 (in Chinese). doi: 10.7524/j.issn.0254-6108.2016.07.2015113001
[2] JING Q F, YI Z L, LIN D H, et al. Enhanced sorption of naphthalene and p-nitrophenol by nano-SiO2 modified with a cationic surfactant[J]. Water Research, 2013, 47(12): 4006-4012. doi: 10.1016/j.watres.2012.09.057 [3] GHOSH A, KHURANA M, CHAUHAN A, et al. Degradation of 4-nitrophenol, 2-chloro-4-nitrophenol, and 2, 4-dinitrophenol by Rhodococcus imtechensis strain RKJ300[J]. Environmental Science & Technology, 2010, 44(3): 1069-1077. [4] 马锋锋, 赵保卫. 玉米芯生物炭吸附水中对硝基苯酚的特性[J]. 环境化学, 2017, 36(4): 898-906. doi: 10.7524/j.issn.0254-6108.2017.04.2016080701 MA F F, ZHAO B W. Adsorption characteristics of p-nitrophenol in aqueous solution by corncob biochar[J]. Environmental Chemistry, 2017, 36(4): 898-906 (in Chinese). doi: 10.7524/j.issn.0254-6108.2017.04.2016080701
[5] 许婉馨, 杨波, 陈子伦, 等. 聚乙烯醇海绵负载铑催化剂催化还原对硝基苯酚[J]. 环境化学, 2020, 39(9): 2576-2583. doi: 10.7524/j.issn.0254-6108.2020050803 XU W X, YANG B, CHEN Z L, et al. Polyvinyl sponge supported RH catalysts for catalytic reduction of P-nitrophenol[J]. Environmental Chemistry, 2020, 39(9): 2576-2583 (in Chinese). doi: 10.7524/j.issn.0254-6108.2020050803
[6] KADAM V V, SHANMUGAM S D, ETTIYAPPAN J P, et al. Photocatalytic degradation of p-nitrophenol using biologically synthesized ZnO nanoparticles[J]. Environmental Science and Pollution Research, 2021, 28(10): 12119-12130. doi: 10.1007/s11356-020-10833-w [7] TANEDA S, MORI Y, KAMATA K, et al. Estrogenic and anti-androgenic activity of nitrophenols in diesel exhaust particles (DEP)[J]. Biological & Pharmaceutical Bulletin, 2004, 27(6): 835-837. [8] LI C M, TANEDA S, SUZUKI A K, et al. Estrogenic and anti-androgenic activities of 4-nitrophenol in diesel exhaust particles[J]. Toxicology and Applied Pharmacology, 2006, 217(1): 1-6. doi: 10.1016/j.taap.2006.06.010 [9] ZHANG Y H, PIAO Y G, LI Y S, et al. 4-Nitrophenol induces Leydig cells hyperplasia, which may contribute to the differential modulation of the androgen receptor and estrogen receptor-α and-β expression in male rat testes[J]. Toxicology Letters, 2013, 223(2): 228-235. doi: 10.1016/j.toxlet.2013.09.011 [10] NI L, YANG C S, GIOELI D, et al. FKBP51 promotes assembly of the Hsp90 chaperone complex and regulates androgen receptor signaling in prostate cancer cells[J]. Molecular and Cellular Biology, 2010, 30(5): 1243-1253. doi: 10.1128/MCB.01891-08 [11] WU D, TAO X Y, CHEN Z P, et al. The environmental endocrine disruptor p-nitrophenol interacts with FKBP51, a positive regulator of androgen receptor and inhibits androgen receptor signaling in human cells[J]. Journal of Hazardous Materials, 2016, 307: 193-201. doi: 10.1016/j.jhazmat.2015.12.045 [12] 李博, 刘书霞, 房森彪, 等. 分子对接与分子动力学计算模拟概论[J]. 比较化学, 2019(1): 1-10. LI B, LIU S X, FANG S B, et al. Progress in molecular docking and molecular dynamics simulation[J]. Journal of Comparative Chemistry, 2019(1): 1-10 (in Chinese).
[13] ZHANG Y, ZENG Z T, ZENG G M, et al. Effect of Triton X-100 on the removal of aqueous phenol by laccase analyzed with a combined approach of experiments and molecular docking[J]. Colloids and Surfaces B: Biointerfaces, 2012, 97: 7-12. doi: 10.1016/j.colsurfb.2012.04.001 [14] DING K K, KONG X T, WANG J P, et al. Side chains of parabens modulate antiandrogenic activity: in vitro and molecular docking studies[J]. Environmental Science & Technology, 2017, 51(11): 6452-6460. [15] NG C A, HUNGERBUEHLER K. Exploring the use of molecular docking to identify bioaccumulative perfluorinated alkyl acids (PFAAs)[J]. Environmental Science & Technology, 2015, 49(20): 12306-12314. [16] 江文婷, 陈旭, 蔡茜茜, 等. 基于分子对接技术研究鱼源抗冻多肽与鱼肌球蛋白的相互作用[J]. 食品工业科技, 2022, 43(20): 29-38. doi: 10.13386/j.issn1002-0306.2022010078 JIANG W T, CHEN X, CAI X X, et al. Prediction of interaction between fish-derived antifreeze peptides and fish myosin by molecular docking[J]. Science and Technology of Food Industry, 2022, 43(20): 29-38 (in Chinese). doi: 10.13386/j.issn1002-0306.2022010078
[17] 赵子晗. CoS2-N/nano-G气体扩散电极电Fenton氧化对硝基苯酚研究[D]. 哈尔滨: 哈尔滨工业大学, 2021. ZHAO Z H. Study on electro-Fenton oxidation of P-nitrophenol by CoS2-N/nano-G gas diffusion electrode[D]. Harbin: Harbin Institute of Technology, 2021 (in Chinese).
[18] YANG J, PAN B, LI H, et al. Degradation of p-nitrophenol on biochars: Role of persistent free radicals[J]. Environmental Science & Technology, 2016, 50(2): 694-700. [19] DELPORT A, HARVEY B H, PETZER A, et al. The monoamine oxidase inhibition properties of selected structural analogues of methylene blue[J]. Toxicology and Applied Pharmacology, 2017, 325: 1-8. doi: 10.1016/j.taap.2017.03.026 [20] RAMSAY R R, DUNFORD C, GILLMAN P K. Methylene blue and serotonin toxicity: Inhibition of monoamine oxidase A (MAO A) confirms a theoretical prediction[J]. British Journal of Pharmacology, 2007, 152(6): 946-951. doi: 10.1038/sj.bjp.0707430 [21] LU C J, ZHOU Q, YAN J, et al. A novel series of tacrine-selegiline hybrids with cholinesterase and monoamine oxidase inhibition activities for the treatment of Alzheimer’s disease[J]. European Journal of Medicinal Chemistry, 2013, 62: 745-753. doi: 10.1016/j.ejmech.2013.01.039 [22] PARK S E, PAUDEL P, WAGLE A, et al. Luteolin, a potent human monoamine oxidase-a inhibitor and dopamine D4 and vasopressin V1A receptor antagonist[J]. Journal of Agricultural and Food Chemistry, 2020, 68(39): 10719-10729. doi: 10.1021/acs.jafc.0c04502 [23] DELPORT A, HARVEY B H, PETZER A, et al. Methylene blue analogues with marginal monoamine oxidase inhibition retain antidepressant-like activity[J]. ACS Chemical Neuroscience, 2018, 9(12): 2917-2928. doi: 10.1021/acschemneuro.8b00042 [24] 袁霞, 徐建业, 吕贞, 等. 卡马西平在UV/氯高级氧化工艺中的去除、转化与毒性评价[J]. 环境化学, 2021, 40(10): 3158-3170. doi: 10.7524/j.issn.0254-6108.2020062701 YUAN X, XU J Y, LYU Z, et al. Removal, transformation and toxicity evaluation of carbamazepine by the UV/chlorine advanced oxidation process [J]. Environmental Chemistry, 2021, 40(10): 3158-3170 (in Chinese). doi: 10.7524/j.issn.0254-6108.2020062701
[25] KUMAR R, MOCHE M, WINBLAD B, et al. Combined X-ray crystallography and computational modeling approach to investigate the Hsp90 C-terminal peptide binding to FKBP51[J]. Scientific Reports, 2017, 7: 14288. doi: 10.1038/s41598-017-14731-z [26] LIU Z F, LIU Y J, ZENG G M, et al. Application of molecular docking for the degradation of organic pollutants in the environmental remediation: A review[J]. Chemosphere, 2018, 203: 139-150. doi: 10.1016/j.chemosphere.2018.03.179 [27] WANG X N, YANG C X, SUN Y Y, et al. A novel screening strategy of anti-SARS-CoV-2 drugs via blocking interaction between Spike RBD and ACE2[J]. Environment International, 2021, 147: 106361. doi: 10.1016/j.envint.2020.106361 [28] YANG C X, WANG X N, ZHANG L L, et al. Investigation of kinetics and mechanism for the degradation of antibiotic norfloxacin in wastewater by UV/H2O2[J]. Journal of the Taiwan Institute of Chemical Engineers, 2020, 115: 117-127. doi: 10.1016/j.jtice.2020.09.036 [29] WANG X N, SUN Y Y, WANG Q, et al. Potential common mechanisms of cytotoxicity induced by amide herbicides via TRPA1 channel activation[J]. International Journal of Environmental Research and Public Health, 2022, 19(13): 7985. doi: 10.3390/ijerph19137985 [30] ZONG W S, WANG X N, DU Y G, et al. Molecular mechanism for the regulation of microcystin toxicity to protein phosphatase 1 by glutathione conjugation pathway[J]. BioMed Research International, 2017, 2017: 9676504. [31] 秦超, 杨兵, 程浩, 等. 多环芳烃与胞外DNA非共价结合及机制[J]. 科学通报, 2022, 67(1): 74-87. doi: 10.1360/TB-2021-0927 QIN C, YANG B, CHENG H, et al. Non-covalent binding interaction and mechanism between polycyclic aromatic hydrocarbons and extracellular DNA[J]. Chinese Science Bulletin, 2022, 67(1): 74-87 (in Chinese). doi: 10.1360/TB-2021-0927
[32] 杨先海, 蔡喜运, 陈景文, 等. 含卤有机化合物与甲状腺素转运蛋白相互作用的卤代效应[J]. 科学通报, 2014, 59(27): 2673-2681. doi: 10.1360/N972013-00059 YANG X H, CAI X Y, CHEN J W, et al. Effects of halogenation on binding interaction between halogenated thyroid disrupting chemicals and transthyretin[J]. Chinese Science Bulletin, 2014, 59(27): 2673-2681 (in Chinese). doi: 10.1360/N972013-00059
-





 下载:
下载: