-
传统生物脱氮的硝化过程可分为两个阶段,第一阶段:氨氧化菌(AOB)将氨氮(NH4+-N)氧化为亚硝酸盐(NO2--N);第二阶段:亚硝酸盐氧化菌(NOB)将亚硝酸盐氧化为硝酸盐(NO3- -N)。因此,硝化反应是由两类微生物独立完成的生化反应过程。
废水中的氨氮因pH值的不同有分子态和离子态两种存在形式。游离氨(FA)是氨氮的分子态形式,通常存在于城市污水和工业废水中。游离氨(FA)浓度可通过Anthonisen 等[1]提出的公式计算得出:
由公式可以看出,FA浓度是氨氮浓度、温度T和pH值三者的函数,并且与三者呈正相关。高氨氮废水的特征是氨氮浓度高,同时还具有较高的pH值,这导致废水中产生较高浓度的FA。高浓度的FA对AOB和NOB的活性具有一定的抑制作用[2-3],降低了生物脱氮硝化过程的生化反应速率,导致系统运行费用增加。几十年来,众多学者针对FA抑制硝化菌的活性方面开展了大量研究[4-11],大多集中于FA选择性抑制 AOB和NOB活性实现短程硝化。此外,FA对AOB和NOB的分解代谢和合成代谢过程均具有一定程度的抑制作用,这揭示了FA抑制其代谢能力的机制[12-14]。
微生物组成是影响生物脱氮效能的关键因素[15]。高通量测序技术是第二代测序技术[16],借助这项技术,许多研究人员研究了生物脱氮硝化过程中FA对细菌种群结构的影响[17-19]。这些研究清楚地表明,随着FA浓度的增加,微生物多样性和硝化细菌丰度均会降低。此外,FA对AOB和NOB活性的抑制阈值的差异[20-22],很可能是由于细菌种群结构的不同引起的。然而,在众多文献中,用于分析微生物群落的活性污泥大多源于FA浓度有差异的生物反应器和污水处理厂,对于在精确/受控条件下驯化的微生物群落的研究较少。为了补充这一研究内容,本试验应用16S r RNA 基因-Illumina MiSeq 高通量测序技术,研究了4种FA浓度(0.5、5、10、15 mg·L−1)条件下长期驯化的活性污泥细菌种群结构的差异以期揭示FA影响生物脱氮硝化过程的生物学机制,为生物脱氮技术的应用提供微生物理论支撑。
-
试验用水采用人工模拟废水,主要成分包括:氯化铵(115 mg·L−1),乙酸钠(385 mg·L−1),磷酸二氢钾(26 mg·L−1)和微量元素浓缩液。接种污泥取自兰州市安宁污水处理厂A2/O工艺好氧段。试验装置为包括SBR反应器(有效容积为4 L)、水浴加热系统及自动控制系统在内(以溶解氧DO、pH值和氧化还原电位ORP为控制参数)的3部分。
-
试验通过控制NH4+ -N浓度,温度和PH值使进水FA浓度分别达到0.5、5、10、15 mg·L−1,对应4个编号为R0.5、R5、R10和R15的SBR反应器。各反应器的溶解氧置于相同水平。具体运行参数见表1。当SBR系统运行稳定时,分别从每个反应器中取3个活性污泥平行样品,共有12个样品。提取所有样品的微生物DNA并进行高通量测序。
-
采用Water DNA Isolation Kit试剂盒提取12个样品中的微生物DNA,将其作为PCR模板, 对16S rRNA 通用引物进行V4 区扩增,在4 ℃下保存。委托上海派森诺生物科技有限公司进行高通量测序。
-
Mothur软件用以计算菌群的Chao1、ACE、Shannon、Simpson多样性指数,RStudio用以获得UniFrac距离矩阵,QIIME2软件用以对样品菌群数据进行流程化分析并生成各分类水平的OTU丰度表。通过Galaxy平台进行LEfSe分析。
-
表2展示了4组样品的α多样性指数。Chao1指数[23]和ACE指数[24]用以评估样本菌群的丰富度,Shannon指数[25]和Simpson指数[26]用以评估样本菌群的均匀度。表2显示4个多样性指数在R0.5中均为最高,而在R15均为最低。这表明R0.5具有最高的微生物多样性,而R15的微生物多样性最低。此外,4组活性污泥样品中序列数和OTU数也基本呈现随FA浓度的增加而逐渐降低的趋势,反映出微生物多样性在R0.5中最高,在R15中最低的特点。
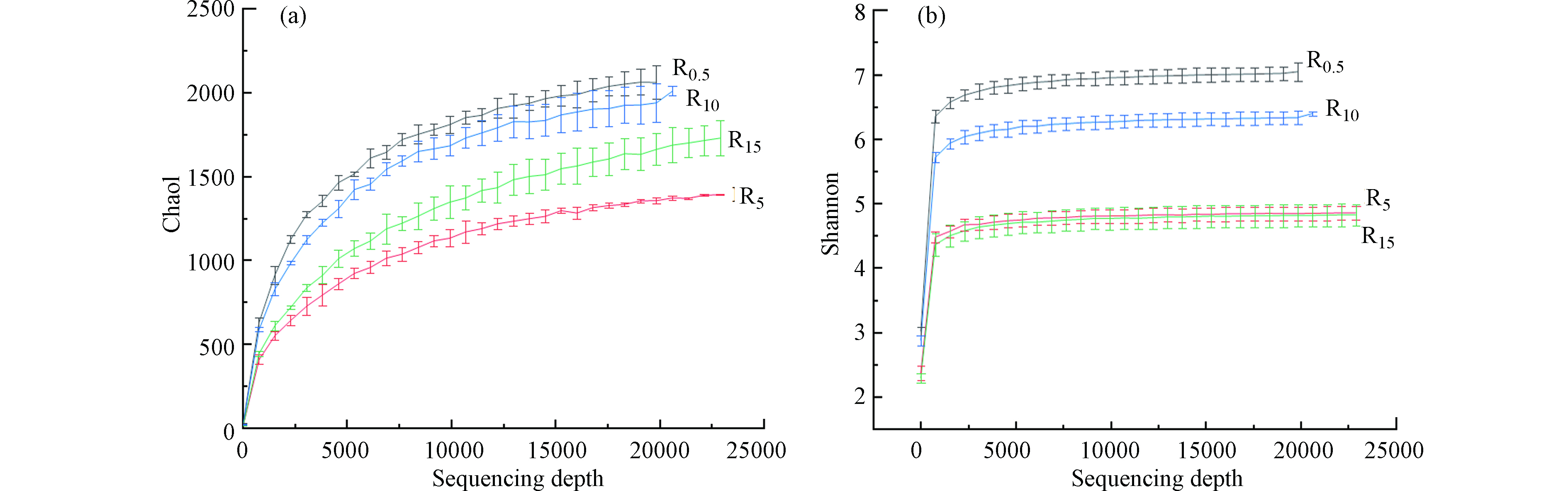
图1为4组活性污泥样品的Chao1和Shannon指数稀疏曲线。随着测序量的深入,Chao1指数稀疏曲线趋于平缓,但还没有饱和。Shannon指数稀疏曲线在后半段非常平缓,说明测序量已足够反映微生物的多样性特征。
-
基于UniFrac距离的非度量多维尺度分析(NMDS)方法用以评估样品微生物β多样性的差异。Unifrac距离分为Unweighted和Weighted两种。Unweighted UniFrac只考虑物种在样品中存在与否 [27];Weighted UniFrac不仅考虑物种的存在与否,还考虑其丰度[28]。
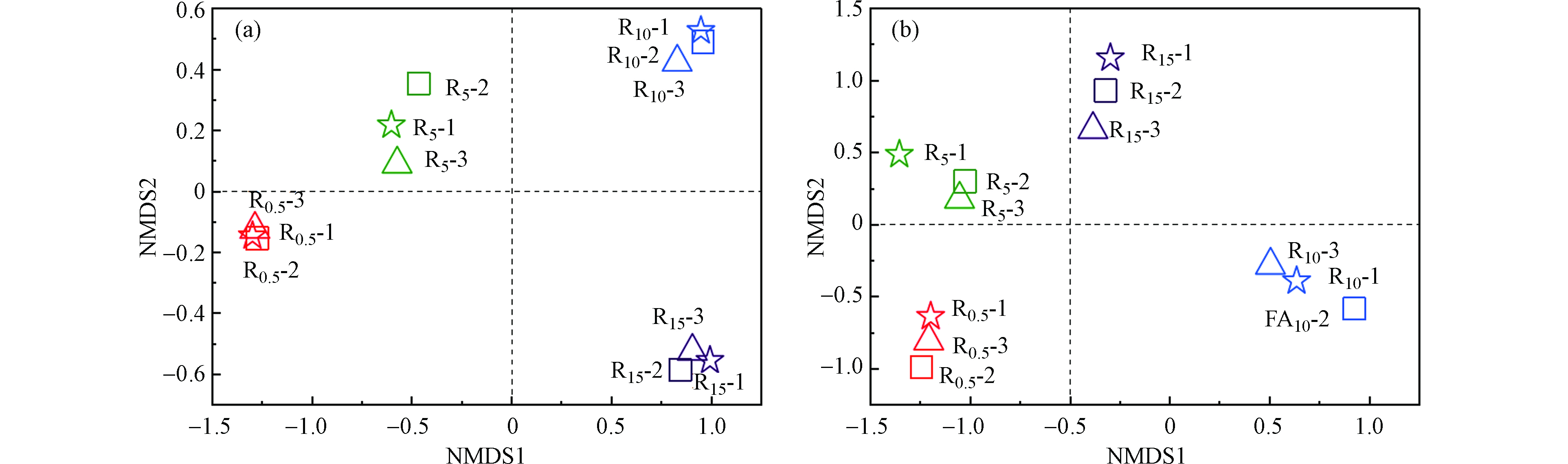
图2为两种基于UniFrac距离的NMDS二维排序图。两两样品之间的距离远近代表样品间菌群结构的相似程度。4组样品组间距离较大,说明不同FA浓度条件下菌群结构差异较大。图2(a)和图2(b)中,R0.5和R5样品的距离较近,说明R0.5和R5的物种组成及丰度较为相似。
-
在门水平上,4组活性污泥样品中共鉴定出32个菌门,其中相对丰度大于0.5%的优势菌门共有9种(表3)。
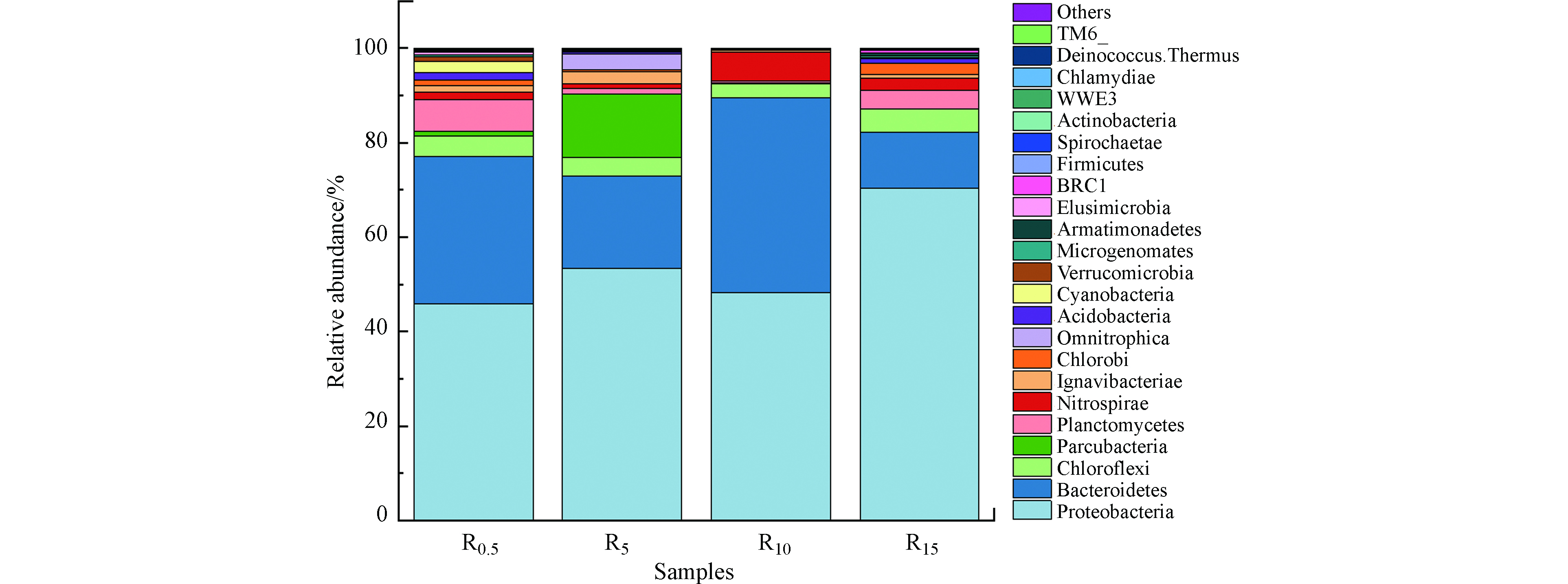
图3为门水平下的细菌种群结构及分布。可以看出,变形菌门(Proteobacteria)、拟杆菌门(Bacteroidetes)、绿弯菌门(Chloroflexi)、Parcubacteria、浮霉菌门(Planctomycetes)、硝化螺旋菌门(Nitrospirae)、Ignavibacteriae、绿菌门(Chlorobi)和Omnitrophica的相对丰度较高,约占总菌门的95.20%—99.62%。这项研究中观察到的细菌种群结构与Cao等[29]的相似, 他利用SBR处理白葡萄酒生产废水。主要区别在于,变形菌门(54.50%)和拟杆菌门(25.95%)的平均相对丰度略低。
FA能够显著影响微生物在门水平上的种群结构和丰度。变形菌门是活性污泥样品中最优势菌门,相对丰度为45.90%—70.46%。众多研究表明,在城市污水处理厂的活性污泥系统中,变形菌门的相对丰度约占27.5%—65%[30],这与本研究获得的结果几乎一致。4 组活性污泥样品中变形菌门的相对丰度均为最高。该菌门在R15(70.46%)的相对丰度最高,在R0.5(45.90%)最低。拟杆菌门的相对丰度在R10(41.25%)最高,在R15(11.84%)最低。此外,一些菌门的相对丰度随着FA浓度的变化表现出独特的变化趋势,如Parcubacteria和Omnitrophica仅在R5中以较高的相对丰度(13.35%和3.37%)存在,而在其它3组活性污泥样品中的相对丰度均低于0.1%。浮霉菌门和绿菌门在R0.5(6.80%和1.24%)和R15(3.96%和2.28%)中具有较高的相对丰度,在R5(1.35%和0.40%)和R10(0.49%和0.03%)的相对丰度较低。Ignavibacteriae在R0.5(1.24%)和R5(2.54%)中均具有较高的相对丰度,在R10(0.38%)和R15(0.89%)的相对丰度较低。需要指出的是,参与硝化反应的主要门类硝化螺旋菌门在R15(0.96%)的相对丰度最低。因此,过高的FA浓度不利于其增殖。
-
在属水平上,4组活性污泥样品中共得到406个菌属。其中陶厄氏菌属(Thauera,31.97%)、腐螺旋菌属(Saprospiraceae,8.78%)、噬纤维菌属(Cytophagaceae,8.38%)、赖文氏菌属(Lewinella,4.86%)和动胶菌属(Zoogloea,4.60%)等相对丰度大于1.0%的菌属共有14种,构成了属水平上的优势菌群(表4)。
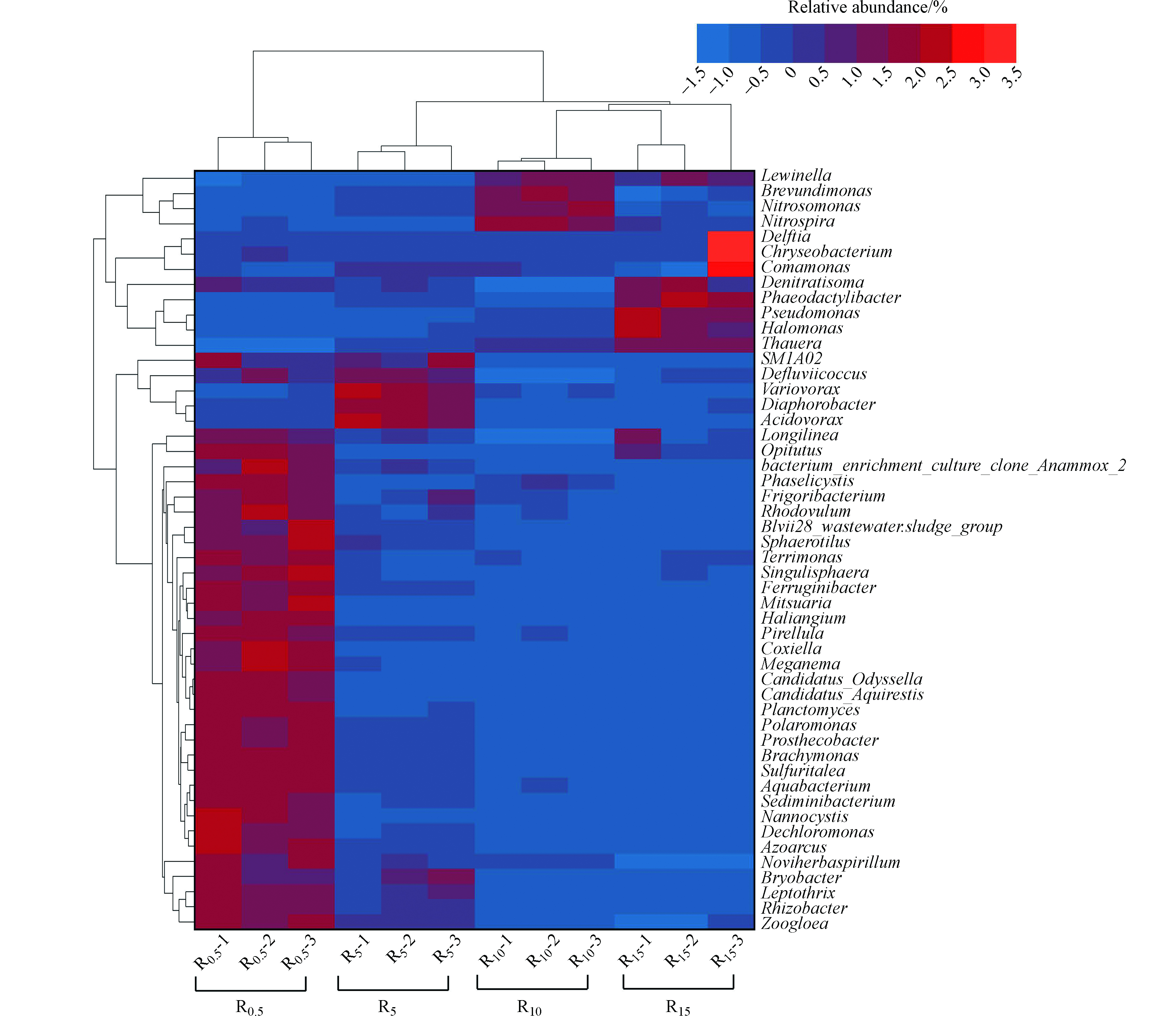
图4为4组样品中丰度前50的属的分层聚类热图,展示了菌属的丰度分布趋势以进一步比较样品间的物种组成差异。基于列聚类(组间和组内)分析,相同FA浓度下平行样品之间的相关性最强,微生物的群落结构组成更相似。列聚类分析表明不同FA浓度下各自形成了特有的菌群。基于行聚类分析,50种菌属可划分为两大分支,进一步划分为4个显著的微生物区域,分别对应4个FA浓度,其中R0.5对应Longilinea—Zoogloea,R5对应SM1A02—Acidovorax,R10对应Lewinella—Nitrospira,R15对应Comamonas—Thauera。
亚硝化单胞菌属(Nitrosomonas)和硝化螺旋菌属(Nitrospira)分别进行氨氮氧化和亚硝酸盐氧化过程[31-33],它们在R10中的丰度最高。需要指出的是,动胶菌属(Zoogloea)和陶厄氏菌属(Thauera)共存于4个SBR系统中,且它们的丰度均较高。此外,这两类菌属的相对丰度与FA浓度之间呈现显著的线性关系。首先,对于动胶菌属,它是污水生物处理中重要的化能异养型兼性好氧菌,对有机物和氮均有一定去除能力,并且是形成菌胶团的主导功能菌属[34-35]。随着FA浓度的增加,动胶菌属的相对丰度逐渐降低,两者呈显著的负相关(y=13.31-3.5x, R2=0.806; Pearson’s ρ=-0.933)。因此,FA浓度增加显著降低了动胶菌属的相对丰度。
对于陶厄氏菌属,它是属于变形菌门的一类革兰氏阴性菌,具有高效的反硝化作用,是污水生物处理中重要的反硝化菌。此外,它还具有降解芳香族化合物的功能[36]。陶厄氏菌属利用污水中的有机物进行新陈代谢活动,在缺氧条件下利用硝态氮作为电子受体进行反硝化,使自身得到增殖[37]。与动胶菌属变化趋势相反,陶厄氏菌属的相对丰度与FA浓度呈显著的正相关(y=-8.72+16.2x, R2=0.97, Pearson’s ρ=0.986)。也就是说,FA对陶厄氏菌属的相对丰度具有显著的促进作用。本研究中,4组样品中陶厄氏菌属的相对丰度(5.05%%—56.29%)远高于Srinandan等 [38]和Du等 [39]的结果。这可能是环境条件和进水质量的差异所致。Cao等的研究中,在FA浓度分别为2.9、5.6、11.1、 19.5 mg·L−1的条件下,陶厄氏菌属(相对丰度< 0.1)并不是优势菌属。这可能是因为其研究中的C / N(4个反应器中的C / N分别为16.5、8.3、4.1和2.4)高于本研究(4个反应器中的C / N为2、0.9、0.6和1.5)。此外,从陶厄氏菌属的生化反应过程来看,在R0.5和R5中,系统硝化结束时硝态氮为主要产物(50 mg·L−1),而在R10和R15中,硝化结束时亚硝态氮为主要产物(30 mg·L−1)。结合陶厄氏菌属相对丰度的变化规律,推断其在还原硝态氮和亚硝态氮的过程中,其更倾向于利用亚硝态氮作为电子受体。
-
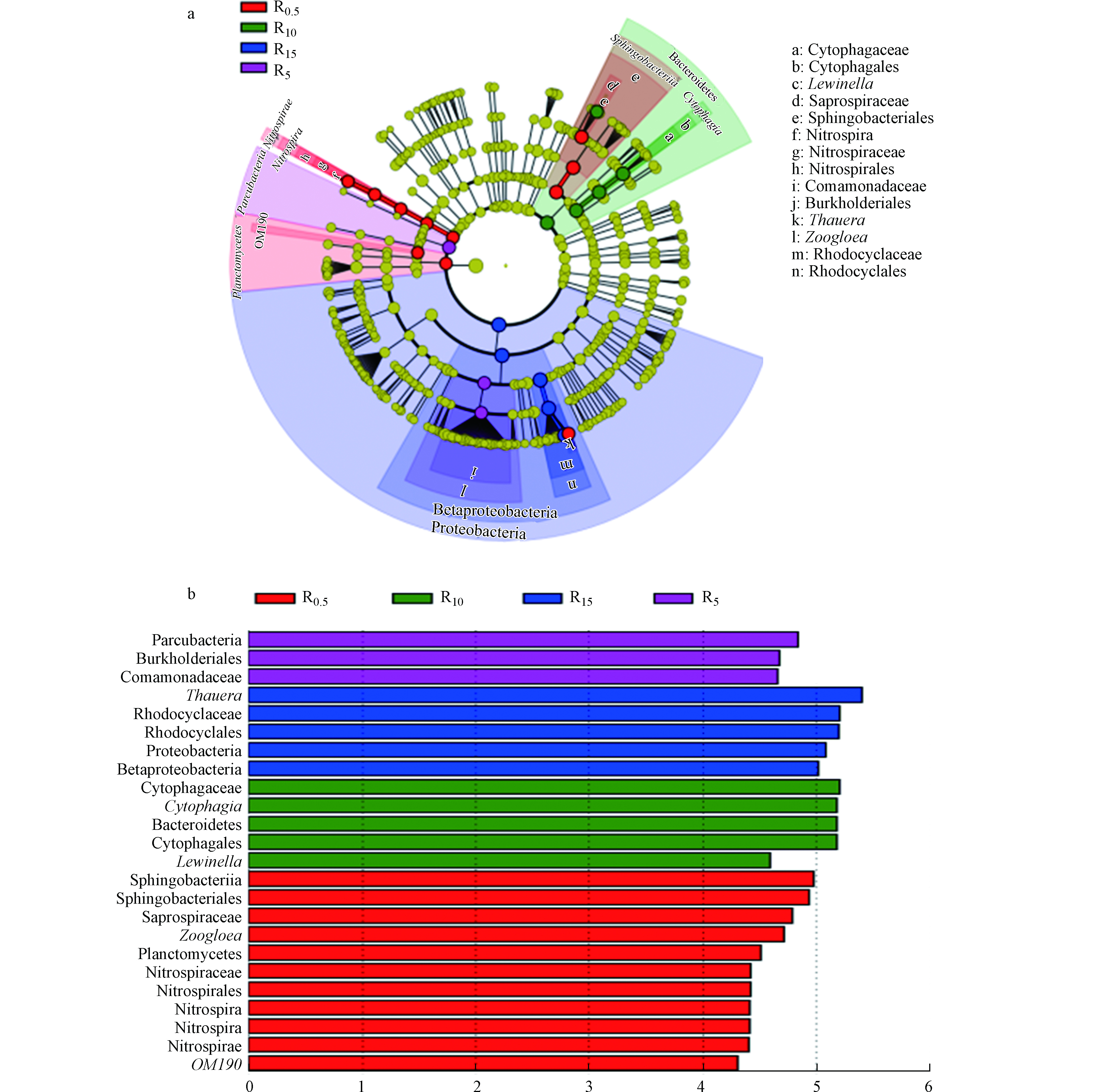
LEfSe(LDA Effect Size)分析方法用以发现4组样品组间在丰度上有显著差异的物种,即微生物标记物。LEfSe分析设定LDA阈值为4.3,P值小于0.05(图5b)。结果表明,共得到24个具有显著差异的微生物标记物(α= 0.01)。R0.5的微生物标记物共有11个,包括鞘脂杆菌纲(Sphingobacteriia)、鞘脂杆菌目(Sphingobacteriales)、腐螺旋菌科(Saprospiraceae)、动胶菌属(Zoogloea)等;R5的微生物标记物共有3个,包括Parcubacteria、伯克氏菌目(Burkholderiales)、丛毛单胞菌科(Comamonadaceae);R10的微生物标记物共有5个,包括噬纤维菌目(Cytophagales)、噬纤维菌属(Cytophagia)、拟杆菌门(Bacteroidetes)、噬纤维菌科(Cytophagaceae)、赖文氏菌属(Lewinella);R15的微生物标记物共有5个,包括陶厄氏菌属(Thauera)、红环菌科(Rhodocyclaceae)、红环菌目(Rhodocyclales)、变形菌门(Proteobacteria)、β-变形菌纲(Betaproteobacteria)。相对丰度最高的微生物标记物分别是R0.5中的鞘脂杆菌纲(28.37%),R5中的Parcubacteria(13.35%),R10中的噬纤维菌目(31.42%)和R15中的陶厄氏菌属(56.29%)。而这4种微生物标志物各自在其他样品中并不显著。从定量的角度来看,4种FA浓度都有利于各自特定的微生物标记物的培养。这表明FA对生物脱氮硝化过程的菌群结构有深刻影响。
-
(1)基于微生物多样性指数,R0.5系统的细菌种群的丰富度和均匀度最高,而R15的细菌种群的丰富度和均匀度最低。FA对系统微生物多样性有显著影响。
(2)在4种FA浓度条件下,细菌种群在门水平上差异显著。最优势菌门变形菌门(Proteobacteria)的相对丰度与FA浓度呈正相关,硝化螺旋菌门(Nitrospirae)的相对丰度与FA浓度呈负相关。拟杆菌门(Bacteroidetes)等其他优势菌门的相对丰度也受FA浓度显著影响。
(3)在4种FA浓度条件下,属水平上形成了不同的优势菌群。亚硝化单胞菌属(Nitrosomonas)和硝化螺旋菌属(Nitrospira)的丰度在R10中丰度明显高于其它3个系统。动胶菌属(Zoogloea)和陶厄氏菌属(Thauera)是两类优势菌属,它们分别与FA浓度呈现显著的负相关和正相关。
(4)LEfSe分析共获得24种具有显著差异的微生物标记物。其中,R0.5中的微生物标记物最多(11个),而R5中的微生物标记物最少(3个)。不同FA浓度条件下,4组样品的关键微生物标记物分别是R0.5中的鞘脂杆菌纲(Sphingobacteriia),R5中的Parcubacteria,R10中的噬纤维菌目(Cytophagales)和R15中的陶厄氏菌属(Thauera)。
游离氨对生物脱氮硝化过程细菌种群结构的影响
Effect of free ammonia on microbial community structure in biological nitrification process
-
摘要: 为揭示游离氨(FA)对硝化过程影响的生物学机制,本试验以人工模拟废水为研究对象,采用4组平行的SBR反应器(R0.5、R5、R10和R15),基于16S rRNA基因-IlluminaMiSeq高通量测序技术,考察了4种FA浓度(0.5、5、10、15 mg·L−1)对SBR反应器中的细菌种群结构的影响。结果表明,FA会显著影响系统内的微生物多样性和菌群结构。R0.5的Chao1、ACE、Shannon和Simpson指数均为最大,其具有最高的微生物多样性,而R15的微生物多样性最低。在微生物门水平上,最优势菌门变形菌门(Proteobacteria)的相对丰度与FA浓度呈正相关,硝化螺旋菌门(Nitrospirae)的相对丰度在R15中最低。在微生物属水平上,亚硝化单胞菌属(Nitrosomonas)和硝化螺旋菌属(Nitrospira)的相对丰度在R10中显著较高,动胶菌属(Zoogloea)和陶厄氏菌属(Thauera)的相对丰度与FA浓度呈显著的线性相关。基于LEfSe分析共获得了24种具有显著差异的微生物,从而得到了4种FA浓度条件下的关键微生物标记物。本研究加深了对生物脱氮硝化过程菌群结构的认识,为深入研究生物脱氮硝化的抑制机理提供了借鉴。
-
关键词:
- 游离氨 /
- Illumina MiSeq高通量测序 /
- 硝化过程 /
- 微生物多样性 /
- LEfSe组间差异
Abstract: Based on 16S rRNA genes-Illumina MiSeq high-throughput sequencing, this study is to investigate the community and structure of characteristic microbial communities related to nitrification under four FA concentrations (0.5, 5, 10, 15 mg·L−1) settled in four parallel laboratory-scale sequencing batch reactors (SBRs, denoted as R0.5, R5, R10, R15) to reveal the biological mechanism of how does FA influence the nitrification. Results indicated that FA concentration significantly affects the microbial diversity and community structure in the system. The largest α-diversity values of Chao1, ACE, Shannon and Simpson were achieved in R0.5, which represents with highest diversity of bacterial community, while the lowest was achieved in R15. At the the phylum level, the relative abundance of Proteobacteria which is the predominant phylum, increased with the increasing of FA concentration. The relative abundance of Nitrospira in the nitrification process was the lowest in R15. At the genus level, the relative abundance of Nitrosomonas and Nitrospira was significantly higher in R10. The relative abundance of Zoogloea and Thauera showed a significant linear correlation with FA concentration. A total of 25 groups of microorganisms with significant differences were obtained based on LEfSe analysis, thereby key biomarkers of the microflora were obtained at the microbiological classification level under four FA concentrations. This study has deepened the understanding of the microbial community structure in the biological nitrification process, and provided a reference for in-depth study of the inhibition mechanism of biological nitrification process. -
传统生物脱氮的硝化过程可分为两个阶段,第一阶段:氨氧化菌(AOB)将氨氮(NH4+-N)氧化为亚硝酸盐(NO2--N);第二阶段:亚硝酸盐氧化菌(NOB)将亚硝酸盐氧化为硝酸盐(NO3- -N)。因此,硝化反应是由两类微生物独立完成的生化反应过程。
废水中的氨氮因pH值的不同有分子态和离子态两种存在形式。游离氨(FA)是氨氮的分子态形式,通常存在于城市污水和工业废水中。游离氨(FA)浓度可通过Anthonisen 等[1]提出的公式计算得出:
FA=1714[NH+4−N]×10pHexp(6334273+T)+10pH (1) 由公式可以看出,FA浓度是氨氮浓度、温度T和pH值三者的函数,并且与三者呈正相关。高氨氮废水的特征是氨氮浓度高,同时还具有较高的pH值,这导致废水中产生较高浓度的FA。高浓度的FA对AOB和NOB的活性具有一定的抑制作用[2-3],降低了生物脱氮硝化过程的生化反应速率,导致系统运行费用增加。几十年来,众多学者针对FA抑制硝化菌的活性方面开展了大量研究[4-11],大多集中于FA选择性抑制 AOB和NOB活性实现短程硝化。此外,FA对AOB和NOB的分解代谢和合成代谢过程均具有一定程度的抑制作用,这揭示了FA抑制其代谢能力的机制[12-14]。
微生物组成是影响生物脱氮效能的关键因素[15]。高通量测序技术是第二代测序技术[16],借助这项技术,许多研究人员研究了生物脱氮硝化过程中FA对细菌种群结构的影响[17-19]。这些研究清楚地表明,随着FA浓度的增加,微生物多样性和硝化细菌丰度均会降低。此外,FA对AOB和NOB活性的抑制阈值的差异[20-22],很可能是由于细菌种群结构的不同引起的。然而,在众多文献中,用于分析微生物群落的活性污泥大多源于FA浓度有差异的生物反应器和污水处理厂,对于在精确/受控条件下驯化的微生物群落的研究较少。为了补充这一研究内容,本试验应用16S r RNA 基因-Illumina MiSeq 高通量测序技术,研究了4种FA浓度(0.5、5、10、15 mg·L−1)条件下长期驯化的活性污泥细菌种群结构的差异以期揭示FA影响生物脱氮硝化过程的生物学机制,为生物脱氮技术的应用提供微生物理论支撑。
1. 材料与方法(Materials and methods)
1.1 试验用水、接种污泥及试验装置
试验用水采用人工模拟废水,主要成分包括:氯化铵(115 mg·L−1),乙酸钠(385 mg·L−1),磷酸二氢钾(26 mg·L−1)和微量元素浓缩液。接种污泥取自兰州市安宁污水处理厂A2/O工艺好氧段。试验装置为包括SBR反应器(有效容积为4 L)、水浴加热系统及自动控制系统在内(以溶解氧DO、pH值和氧化还原电位ORP为控制参数)的3部分。
1.2 试验方案
试验通过控制NH4+ -N浓度,温度和PH值使进水FA浓度分别达到0.5、5、10、15 mg·L−1,对应4个编号为R0.5、R5、R10和R15的SBR反应器。各反应器的溶解氧置于相同水平。具体运行参数见表1。当SBR系统运行稳定时,分别从每个反应器中取3个活性污泥平行样品,共有12个样品。提取所有样品的微生物DNA并进行高通量测序。
表 1 SBR反应器运行条件Table 1. Operating conditions of SBR reactorsFA/(mg·L−1) 初始运行参数Initial operational parameters 运行周期及各阶段反应时间/minPhase time of the SBR COD/(mg·L−1) NH4+-N /(mg·L−1) 温度/ ℃Temperature pH 周期时间One cycle 进水Filling 曝气Aeration 缺氧Anoxia 沉淀排水Setting 0.5 80 40 20±2.0 7.5±0.2 520 5 270 180 45 5 80 90 25±2.0 8.0±0.2 570 5 300 200 25 10 80 130 30±2.0 8.0±0.2 660 5 360 240 25 15 80 55 35±2.0 8.5±0.2 490 5 240 160 45 1.3 高通量测序
采用Water DNA Isolation Kit试剂盒提取12个样品中的微生物DNA,将其作为PCR模板, 对16S rRNA 通用引物进行V4 区扩增,在4 ℃下保存。委托上海派森诺生物科技有限公司进行高通量测序。
1.4 菌群多样性和差异性分析
Mothur软件用以计算菌群的Chao1、ACE、Shannon、Simpson多样性指数,RStudio用以获得UniFrac距离矩阵,QIIME2软件用以对样品菌群数据进行流程化分析并生成各分类水平的OTU丰度表。通过Galaxy平台进行LEfSe分析。
2. 结果与讨论(Results and discussion)
2.1 FA对系统菌群多样性的影响
2.1.1 FA对菌群α多样性的影响
表2展示了4组样品的α多样性指数。Chao1指数[23]和ACE指数[24]用以评估样本菌群的丰富度,Shannon指数[25]和Simpson指数[26]用以评估样本菌群的均匀度。表2显示4个多样性指数在R0.5中均为最高,而在R15均为最低。这表明R0.5具有最高的微生物多样性,而R15的微生物多样性最低。此外,4组活性污泥样品中序列数和OTU数也基本呈现随FA浓度的增加而逐渐降低的趋势,反映出微生物多样性在R0.5中最高,在R15中最低的特点。
表 2 4组活性污泥样品中微生物群落的丰富度及多样性Table 2. Four groups in activated sludge samples the richness and diversity of microbial communities样品Samples 有效序列数Amount of effective sequencing OTUs 微生物数量Amount of microbial groups 细菌群落的α多样性α-diversity of bacterial community 门Phylum 纲Class 目Order 科Family 属Genus 种Species Chao1 ACE Shannon Simpson R0.5 45637 2766 27 55 69 101 137 54 1915 1915 8.83 0.99 R5 40901 2521 20 48 58 79 107 47 1731 1732 7.85 0.97 R10 44160 1349 17 31 44 57 68 33 1238 1268 6.24 0.94 R15 38423 1438 20 39 59 74 79 41 1169 1179 5.68 0.86 图1为4组活性污泥样品的Chao1和Shannon指数稀疏曲线。随着测序量的深入,Chao1指数稀疏曲线趋于平缓,但还没有饱和。Shannon指数稀疏曲线在后半段非常平缓,说明测序量已足够反映微生物的多样性特征。
2.1.2 FA对菌群β多样性的影响
基于UniFrac距离的非度量多维尺度分析(NMDS)方法用以评估样品微生物β多样性的差异。Unifrac距离分为Unweighted和Weighted两种。Unweighted UniFrac只考虑物种在样品中存在与否 [27];Weighted UniFrac不仅考虑物种的存在与否,还考虑其丰度[28]。
图2为两种基于UniFrac距离的NMDS二维排序图。两两样品之间的距离远近代表样品间菌群结构的相似程度。4组样品组间距离较大,说明不同FA浓度条件下菌群结构差异较大。图2(a)和图2(b)中,R0.5和R5样品的距离较近,说明R0.5和R5的物种组成及丰度较为相似。
2.2 FA对系统菌群组成的影响
2.2.1 门水平上的菌群分布特征
在门水平上,4组活性污泥样品中共鉴定出32个菌门,其中相对丰度大于0.5%的优势菌门共有9种(表3)。
表 3 门水平上样品中的主要菌群Table 3. The main flora in the samples at the phylum level门水平微生物Taxon 总计Total 样品Samples R0.5 R5 R10 R15 变形菌门(Proteobacteria) 54.50% 45.90% 53.49% 48.17% 70.46% 拟杆菌门(Bacteroidetes) 25.95% 31.17% 19.55% 41.25% 11.84% 绿弯菌门(Chloroflexi) 4.05% 4.40% 3.85% 3.10% 4.86% Parcubacteria 3.59% 0.95% 13.35% 0.06% 0.01% 浮霉菌门(Planctomycetes) 3.15% 6.80% 1.35% 0.49% 3.96% 硝化螺旋菌门(Nitrospirae) 2.78% 6.15% 2.44% 1.57% 0.96% Ignavibacteriae 1.27% 1.24% 2.54% 0.38% 0.89% 绿菌门(Chlorobi) 0.99% 1.24% 0.40% 0.03% 2.28% Omnitrophica 0.60% 0.01% 2.37% 0.00% 0.00% 图3为门水平下的细菌种群结构及分布。可以看出,变形菌门(Proteobacteria)、拟杆菌门(Bacteroidetes)、绿弯菌门(Chloroflexi)、Parcubacteria、浮霉菌门(Planctomycetes)、硝化螺旋菌门(Nitrospirae)、Ignavibacteriae、绿菌门(Chlorobi)和Omnitrophica的相对丰度较高,约占总菌门的95.20%—99.62%。这项研究中观察到的细菌种群结构与Cao等[29]的相似, 他利用SBR处理白葡萄酒生产废水。主要区别在于,变形菌门(54.50%)和拟杆菌门(25.95%)的平均相对丰度略低。
FA能够显著影响微生物在门水平上的种群结构和丰度。变形菌门是活性污泥样品中最优势菌门,相对丰度为45.90%—70.46%。众多研究表明,在城市污水处理厂的活性污泥系统中,变形菌门的相对丰度约占27.5%—65%[30],这与本研究获得的结果几乎一致。4 组活性污泥样品中变形菌门的相对丰度均为最高。该菌门在R15(70.46%)的相对丰度最高,在R0.5(45.90%)最低。拟杆菌门的相对丰度在R10(41.25%)最高,在R15(11.84%)最低。此外,一些菌门的相对丰度随着FA浓度的变化表现出独特的变化趋势,如Parcubacteria和Omnitrophica仅在R5中以较高的相对丰度(13.35%和3.37%)存在,而在其它3组活性污泥样品中的相对丰度均低于0.1%。浮霉菌门和绿菌门在R0.5(6.80%和1.24%)和R15(3.96%和2.28%)中具有较高的相对丰度,在R5(1.35%和0.40%)和R10(0.49%和0.03%)的相对丰度较低。Ignavibacteriae在R0.5(1.24%)和R5(2.54%)中均具有较高的相对丰度,在R10(0.38%)和R15(0.89%)的相对丰度较低。需要指出的是,参与硝化反应的主要门类硝化螺旋菌门在R15(0.96%)的相对丰度最低。因此,过高的FA浓度不利于其增殖。
2.2.2 属水平上的菌群分布特征
在属水平上,4组活性污泥样品中共得到406个菌属。其中陶厄氏菌属(Thauera,31.97%)、腐螺旋菌属(Saprospiraceae,8.78%)、噬纤维菌属(Cytophagaceae,8.38%)、赖文氏菌属(Lewinella,4.86%)和动胶菌属(Zoogloea,4.60%)等相对丰度大于1.0%的菌属共有14种,构成了属水平上的优势菌群(表4)。
表 4 属水平上样品中的主要菌群Table 4. The main flora in the samples at the genus level属水平微生物 Taxon 总计 Total 样品Samples R0.5 R5 R10 R15 陶厄氏菌属(Thauera) 31.97% 5.05% 28.76% 37.78% 56.29% 腐螺旋菌属(Saprospiraceae) 8.78% 21.45% 13.08% 0.36% 0.24% 噬纤维菌属(Cytophagaceae) 8.38% 1.46% 0.09% 31.10% 0.85% 赖文氏菌属(Lewinella) 4.86% 1.21% 1.44% 9.03% 7.76% 动胶菌属(Zoogloea) 4.60% 11.23% 5.12% 1.09% 0.94% 厌氧绳菌属(Anaerolineaceae) 3.21% 3.28% 3.16% 2.22% 4.16% 硝化螺旋菌属(Nitrospira) 2.78% 1.57% 0.96% 6.14% 2.44% 脱氯菌属(Dechloromonas) 2.40% 0.04% 9.51% 0.05% 0.00% 丛毛单胞菌属(Comamonadaceae) 1.63% 2.66% 2.32% 1.20% 0.33% OM190 1.42% 4.46% 0.88% 0.30% 0.05% 固氮弓菌属(Azoarcus) 1.30% 3.68% 0.85% 0.38% 0.30% 食酸菌属(Acidovorax) 1.27% 1.08% 3.88% 0.09% 0.03% 亚硝化单胞菌属(Nitrosomonas) 1.25% 0.13% 1.09% 3.17% 0.60% 浮霉菌属(Planctomycetaceae) 1.01% 0.12% 0.11% 0.07% 3.76% 图4为4组样品中丰度前50的属的分层聚类热图,展示了菌属的丰度分布趋势以进一步比较样品间的物种组成差异。基于列聚类(组间和组内)分析,相同FA浓度下平行样品之间的相关性最强,微生物的群落结构组成更相似。列聚类分析表明不同FA浓度下各自形成了特有的菌群。基于行聚类分析,50种菌属可划分为两大分支,进一步划分为4个显著的微生物区域,分别对应4个FA浓度,其中R0.5对应Longilinea—Zoogloea,R5对应SM1A02—Acidovorax,R10对应Lewinella—Nitrospira,R15对应Comamonas—Thauera。
亚硝化单胞菌属(Nitrosomonas)和硝化螺旋菌属(Nitrospira)分别进行氨氮氧化和亚硝酸盐氧化过程[31-33],它们在R10中的丰度最高。需要指出的是,动胶菌属(Zoogloea)和陶厄氏菌属(Thauera)共存于4个SBR系统中,且它们的丰度均较高。此外,这两类菌属的相对丰度与FA浓度之间呈现显著的线性关系。首先,对于动胶菌属,它是污水生物处理中重要的化能异养型兼性好氧菌,对有机物和氮均有一定去除能力,并且是形成菌胶团的主导功能菌属[34-35]。随着FA浓度的增加,动胶菌属的相对丰度逐渐降低,两者呈显著的负相关(y=13.31-3.5x, R2=0.806; Pearson’s ρ=-0.933)。因此,FA浓度增加显著降低了动胶菌属的相对丰度。
对于陶厄氏菌属,它是属于变形菌门的一类革兰氏阴性菌,具有高效的反硝化作用,是污水生物处理中重要的反硝化菌。此外,它还具有降解芳香族化合物的功能[36]。陶厄氏菌属利用污水中的有机物进行新陈代谢活动,在缺氧条件下利用硝态氮作为电子受体进行反硝化,使自身得到增殖[37]。与动胶菌属变化趋势相反,陶厄氏菌属的相对丰度与FA浓度呈显著的正相关(y=-8.72+16.2x, R2=0.97, Pearson’s ρ=0.986)。也就是说,FA对陶厄氏菌属的相对丰度具有显著的促进作用。本研究中,4组样品中陶厄氏菌属的相对丰度(5.05%%—56.29%)远高于Srinandan等 [38]和Du等 [39]的结果。这可能是环境条件和进水质量的差异所致。Cao等的研究中,在FA浓度分别为2.9、5.6、11.1、 19.5 mg·L−1的条件下,陶厄氏菌属(相对丰度< 0.1)并不是优势菌属。这可能是因为其研究中的C / N(4个反应器中的C / N分别为16.5、8.3、4.1和2.4)高于本研究(4个反应器中的C / N为2、0.9、0.6和1.5)。此外,从陶厄氏菌属的生化反应过程来看,在R0.5和R5中,系统硝化结束时硝态氮为主要产物(50 mg·L−1),而在R10和R15中,硝化结束时亚硝态氮为主要产物(30 mg·L−1)。结合陶厄氏菌属相对丰度的变化规律,推断其在还原硝态氮和亚硝态氮的过程中,其更倾向于利用亚硝态氮作为电子受体。
2.3 不同FA浓度样品的微生物标记物
LEfSe(LDA Effect Size)分析方法用以发现4组样品组间在丰度上有显著差异的物种,即微生物标记物。LEfSe分析设定LDA阈值为4.3,P值小于0.05(图5b)。结果表明,共得到24个具有显著差异的微生物标记物(α= 0.01)。R0.5的微生物标记物共有11个,包括鞘脂杆菌纲(Sphingobacteriia)、鞘脂杆菌目(Sphingobacteriales)、腐螺旋菌科(Saprospiraceae)、动胶菌属(Zoogloea)等;R5的微生物标记物共有3个,包括Parcubacteria、伯克氏菌目(Burkholderiales)、丛毛单胞菌科(Comamonadaceae);R10的微生物标记物共有5个,包括噬纤维菌目(Cytophagales)、噬纤维菌属(Cytophagia)、拟杆菌门(Bacteroidetes)、噬纤维菌科(Cytophagaceae)、赖文氏菌属(Lewinella);R15的微生物标记物共有5个,包括陶厄氏菌属(Thauera)、红环菌科(Rhodocyclaceae)、红环菌目(Rhodocyclales)、变形菌门(Proteobacteria)、β-变形菌纲(Betaproteobacteria)。相对丰度最高的微生物标记物分别是R0.5中的鞘脂杆菌纲(28.37%),R5中的Parcubacteria(13.35%),R10中的噬纤维菌目(31.42%)和R15中的陶厄氏菌属(56.29%)。而这4种微生物标志物各自在其他样品中并不显著。从定量的角度来看,4种FA浓度都有利于各自特定的微生物标记物的培养。这表明FA对生物脱氮硝化过程的菌群结构有深刻影响。
3. 结论(Conclusion)
(1)基于微生物多样性指数,R0.5系统的细菌种群的丰富度和均匀度最高,而R15的细菌种群的丰富度和均匀度最低。FA对系统微生物多样性有显著影响。
(2)在4种FA浓度条件下,细菌种群在门水平上差异显著。最优势菌门变形菌门(Proteobacteria)的相对丰度与FA浓度呈正相关,硝化螺旋菌门(Nitrospirae)的相对丰度与FA浓度呈负相关。拟杆菌门(Bacteroidetes)等其他优势菌门的相对丰度也受FA浓度显著影响。
(3)在4种FA浓度条件下,属水平上形成了不同的优势菌群。亚硝化单胞菌属(Nitrosomonas)和硝化螺旋菌属(Nitrospira)的丰度在R10中丰度明显高于其它3个系统。动胶菌属(Zoogloea)和陶厄氏菌属(Thauera)是两类优势菌属,它们分别与FA浓度呈现显著的负相关和正相关。
(4)LEfSe分析共获得24种具有显著差异的微生物标记物。其中,R0.5中的微生物标记物最多(11个),而R5中的微生物标记物最少(3个)。不同FA浓度条件下,4组样品的关键微生物标记物分别是R0.5中的鞘脂杆菌纲(Sphingobacteriia),R5中的Parcubacteria,R10中的噬纤维菌目(Cytophagales)和R15中的陶厄氏菌属(Thauera)。
-
表 1 SBR反应器运行条件
Table 1. Operating conditions of SBR reactors
FA/(mg·L−1) 初始运行参数Initial operational parameters 运行周期及各阶段反应时间/minPhase time of the SBR COD/(mg·L−1) NH4+-N /(mg·L−1) 温度/ ℃Temperature pH 周期时间One cycle 进水Filling 曝气Aeration 缺氧Anoxia 沉淀排水Setting 0.5 80 40 20±2.0 7.5±0.2 520 5 270 180 45 5 80 90 25±2.0 8.0±0.2 570 5 300 200 25 10 80 130 30±2.0 8.0±0.2 660 5 360 240 25 15 80 55 35±2.0 8.5±0.2 490 5 240 160 45 表 2 4组活性污泥样品中微生物群落的丰富度及多样性
Table 2. Four groups in activated sludge samples the richness and diversity of microbial communities
样品Samples 有效序列数Amount of effective sequencing OTUs 微生物数量Amount of microbial groups 细菌群落的α多样性α-diversity of bacterial community 门Phylum 纲Class 目Order 科Family 属Genus 种Species Chao1 ACE Shannon Simpson R0.5 45637 2766 27 55 69 101 137 54 1915 1915 8.83 0.99 R5 40901 2521 20 48 58 79 107 47 1731 1732 7.85 0.97 R10 44160 1349 17 31 44 57 68 33 1238 1268 6.24 0.94 R15 38423 1438 20 39 59 74 79 41 1169 1179 5.68 0.86 表 3 门水平上样品中的主要菌群
Table 3. The main flora in the samples at the phylum level
门水平微生物Taxon 总计Total 样品Samples R0.5 R5 R10 R15 变形菌门(Proteobacteria) 54.50% 45.90% 53.49% 48.17% 70.46% 拟杆菌门(Bacteroidetes) 25.95% 31.17% 19.55% 41.25% 11.84% 绿弯菌门(Chloroflexi) 4.05% 4.40% 3.85% 3.10% 4.86% Parcubacteria 3.59% 0.95% 13.35% 0.06% 0.01% 浮霉菌门(Planctomycetes) 3.15% 6.80% 1.35% 0.49% 3.96% 硝化螺旋菌门(Nitrospirae) 2.78% 6.15% 2.44% 1.57% 0.96% Ignavibacteriae 1.27% 1.24% 2.54% 0.38% 0.89% 绿菌门(Chlorobi) 0.99% 1.24% 0.40% 0.03% 2.28% Omnitrophica 0.60% 0.01% 2.37% 0.00% 0.00% 表 4 属水平上样品中的主要菌群
Table 4. The main flora in the samples at the genus level
属水平微生物 Taxon 总计 Total 样品Samples R0.5 R5 R10 R15 陶厄氏菌属(Thauera) 31.97% 5.05% 28.76% 37.78% 56.29% 腐螺旋菌属(Saprospiraceae) 8.78% 21.45% 13.08% 0.36% 0.24% 噬纤维菌属(Cytophagaceae) 8.38% 1.46% 0.09% 31.10% 0.85% 赖文氏菌属(Lewinella) 4.86% 1.21% 1.44% 9.03% 7.76% 动胶菌属(Zoogloea) 4.60% 11.23% 5.12% 1.09% 0.94% 厌氧绳菌属(Anaerolineaceae) 3.21% 3.28% 3.16% 2.22% 4.16% 硝化螺旋菌属(Nitrospira) 2.78% 1.57% 0.96% 6.14% 2.44% 脱氯菌属(Dechloromonas) 2.40% 0.04% 9.51% 0.05% 0.00% 丛毛单胞菌属(Comamonadaceae) 1.63% 2.66% 2.32% 1.20% 0.33% OM190 1.42% 4.46% 0.88% 0.30% 0.05% 固氮弓菌属(Azoarcus) 1.30% 3.68% 0.85% 0.38% 0.30% 食酸菌属(Acidovorax) 1.27% 1.08% 3.88% 0.09% 0.03% 亚硝化单胞菌属(Nitrosomonas) 1.25% 0.13% 1.09% 3.17% 0.60% 浮霉菌属(Planctomycetaceae) 1.01% 0.12% 0.11% 0.07% 3.76% -
[1] ANTHONISEN A C, LOEHR R C, PRAKASAM T B S, et al. Inhibition of nitrification by ammonia and nitrousacid [J]. Journal Water Pollution Control Federation, 1976, 48(5): 835-52. [2] FUX C, BOEHLER M, HUBER P, et al. Biological treatment of ammonium-rich wastewater by partial nitritation and subsequent anaerobic ammonium oxidation (anammox) in a pilot plant [J]. Journal of Biotechnology, 2002, 99(3): 295-306. doi: 10.1016/S0168-1656(02)00220-1 [3] KIM D J, LEE D I, KELLER J. Effect of temperature and free ammonia on nitrification and nitrite accumulation in landfill leachate and analysis of its nitrifying bacterial community by FISH [J]. Bioresource Technology, 2006, 97(3): 459-468. doi: 10.1016/j.biortech.2005.03.032 [4] JIANG Y S, POH L S, LIM C P, et al. Effect of free ammonia inhibition on process recovery of partial nitritation in a membrane bioreactor [J]. Bioresource Technology Reports, 2019, 6: 152-158. doi: 10.1016/j.biteb.2019.02.014 [5] 孙洪伟, 尤永军, 赵华南, 等. 游离氨对硝化菌活性的抑制及可逆性影响 [J]. 中国环境科学, 2015, 35(1): 95-100. SUN H W, YOU Y J, ZHAO H N, et al. Inhibitory effect of free ammonia on the activity of nitrifying bacteria and recoverability [J]. China Environmental Science, 2015, 35(1): 95-100(in Chinese).
[6] 孙洪伟, 吕心涛, 魏雪芬, 等. 游离氨(FA)耦合曝气时间对硝化菌活性的抑制影响 [J]. 环境科学, 2016, 37(3): 1075-1081. SUN H W, LÜ X T, WEI X F, et al. Synergetic inhibitory effect of free ammonia and aeration phase length control on the activity of nitrifying bacteria [J]. Environmental Science, 2016, 37(3): 1075-1081(in Chinese).
[7] 张亮, 张树军, 彭永臻. 污水处理中游离氨对硝化作用抑制影响研究 [J]. 哈尔滨工业大学学报, 2012, 44(2): 75-79. doi: 10.11918/j.issn.0367-6234.2012.02.016 ZHANG L, ZHANG S J, PENG Y Z. Review of study on the effects of free ammonia inhibitions on nitrifying bacteria in wastewater treatment [J]. Journal of Harbin Institute of Technology, 2012, 44(2): 75-79(in Chinese). doi: 10.11918/j.issn.0367-6234.2012.02.016
[8] YAO Q, PENG D C, WANG B, et al. Effect of free ammonium and free nitrous acid on the activity, aggregate morphology and extracellular polymeric substance distribution of ammonium oxidizing bacteria in partial nitrification [J]. Journal of Bioscience and Bioengineering, 2017, 124(3): 319-326. doi: 10.1016/j.jbiosc.2017.03.015 [9] POOT V, HOEKSTRA M, GELEIJNSE M A A, et al. Effects of the residual ammonium concentration on NOB repression during partial nitritation with granular sludge [J]. Water Research, 2016, 106: 518-530. doi: 10.1016/j.watres.2016.10.028 [10] KENT T R, SUN Y W, AN Z H, et al. Mechanistic understanding of the NOB suppression by free ammonia inhibition in continuous flow aerobic granulation bioreactors [J]. Environment International, 2019, 131: 105005. doi: 10.1016/j.envint.2019.105005 [11] DUAN H R, YE L, WANG Q L, et al. Nitrite oxidizing bacteria (NOB) contained in influent deteriorate mainstream NOB suppression by sidestream inactivation [J]. Water Research, 2019, 162: 331-338. doi: 10.1016/j.watres.2019.07.002 [12] LIU Y W, NGO H H, GUO W S, et al. The roles of free ammonia (FA) in biological wastewater treatment processes: A review [J]. Environment International, 2019, 123: 10-19. doi: 10.1016/j.envint.2018.11.039 [13] WANG B, WANG Z H, WANG S Y, et al. Recovering partial nitritation in a PN/A system during mainstream wastewater treatment by reviving AOB activity after thoroughly inhibiting AOB and NOB with free nitrous acid [J]. Environment International, 2020, 139: 105684. doi: 10.1016/j.envint.2020.105684 [14] YU L T, CHEN S D, CHEN W J, et al. Experimental investigation and mathematical modeling of the competition among the fast-growing “r-strategists” and the slow-growing “K-strategists” ammonium-oxidizing bacteria and nitrite-oxidizing bacteria in nitrification [J]. Science of the Total Environment, 2020, 702: 135049. doi: 10.1016/j.scitotenv.2019.135049 [15] ZHANG Q H, YANG W N, NGO H H, et al. Current status of urban wastewater treatment plants in China [J]. Environment International, 2016, 92/93: 11-22. doi: 10.1016/j.envint.2016.03.024 [16] CHU Y J, COREY D R. RNA sequencing: Platform selection, experimental design, and data interpretation [J]. Nucleic Acid Therapeutics, 2012, 22(4): 271-274. doi: 10.1089/nat.2012.0367 [17] VADIVELU V M, KELLER J, YUAN Z G. Effect of free ammonia and free nitrous acid concentration on the anabolic and catabolic processes of an enriched Nitrosomonas culture [J]. Biotechnology and Bioengineering, 2006, 95(5): 830-839. doi: 10.1002/bit.21018 [18] CHEN W J, DAI X H, CAO D W, et al. Performance and microbial ecology of a nitritation sequencing batch reactor treating high-strength ammonia wastewater [J]. Scientific Reports, 2016, 6: 35693. doi: 10.1038/srep35693 [19] SUI Q W, LIU C, ZHANG J Y, et al. Response of nitrite accumulation and microbial community to free ammonia and dissolved oxygen treatment of high ammonium wastewater [J]. Applied Microbiology and Biotechnology, 2016, 100(9): 4177-4187. doi: 10.1007/s00253-015-7183-z [20] ZHANG C, QIN Y G, XU Q X, et al. Free ammonia-based pretreatment promotes short-chain fatty acid production from waste activated sludge [J]. ACS Sustainable Chemistry & Engineering, 2018, 6(7): 9120-9129. [21] VADIVELU V M, KELLER J, YUAN Z G. Effect of free ammonia on the respiration and growth processes of an enriched Nitrobacter culture [J]. Water Research, 2007, 41(4): 826-834. doi: 10.1016/j.watres.2006.11.030 [22] HULLE S W, VOLCKE E I, TERUEL J L, et al. Influence of temperature and pH on the kinetics of the Sharon nitritation process [J]. Journal of Chemical Technology & Biotechnology, 2007, 82(5): 471-480. [23] CHAO A. Nonparametric estimation of the number of classes in a population [J]. Scand J Statist, 1984, 11(4): 265-270. [24] CHAO A, YANG M C K. Stopping rules and estimation for recapture debugging with unequal failure rates [J]. Biometrika, 1993, 80(1): 193-201. doi: 10.1093/biomet/80.1.193 [25] SHANNON C E. A mathematical theory of communication [J]. Bell System Technical Journal, 1948, 27(3): 379-423. doi: 10.1002/j.1538-7305.1948.tb01338.x [26] SIMPSON E H. Measurement of diversity [J]. Journal of Cardiothoracic & Vascular Anesthesia, 1997, 11(6): 812-812. [27] FANG D X, ZHAO G, XU X Y, et al. Microbial community structures and functions of wastewater treatment systems in plateau and cold regions [J]. Bioresource Technology, 2018, 249: 684-693. doi: 10.1016/j.biortech.2017.10.063 [28] USHIKI N, FUJITANI H, AOI Y, et al. Isolation of Nitrospira belonging to sublineage II from a wastewater treatment plant [J]. Microbes and Environments, 2013, 28(3): 346-353. doi: 10.1264/jsme2.ME13042 [29] CAO J S, YU Y X, XIE K, et al. Characterizing the free ammonia exposure to the nutrients removal in activated sludge systems [J]. RSC Advances, 2017, 7(87): 55088-55097. doi: 10.1039/C7RA10751J [30] GILBERT E M, AGRAWAL S, BRUNNER F, et al. Response of different Nitrospira species to anoxic periods depends on operational DO [J]. Environmental Science & Technology, 2014, 48(5): 2934-2941. [31] CHIELLINI C, MUNZ G, PETRONI G, et al. Characterization and comparison of bacterial communities selected in conventional activated sludge and membrane bioreactor pilot plants: A focus on Nitrospira and planctomycetes bacterial Phyla [J]. Current Microbiology, 2013, 67(1): 77-90. doi: 10.1007/s00284-013-0333-6 [32] LOZUPONE C A, HAMADY M, KELLEY S T, et al. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities [J]. Applied and Environmental Microbiology, 2007, 73(5): 1576-1585. doi: 10.1128/AEM.01996-06 [33] ZHANG T, SHAO M F, YE L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants [J]. The ISME Journal, 2012, 6(6): 1137-1147. doi: 10.1038/ismej.2011.188 [34] SUN H W, SHI W Y, CAI C J, et al. Responses of microbial structures, functions, metabolic pathways and community interactions to different C/N ratios in aerobic nitrification [J]. Bioresource Technology, 2020, 311: 123422. doi: 10.1016/j.biortech.2020.123422 [35] CHEN H, LI A, CUI D, et al. Evolution of microbial community and key genera in the formation and stability of aerobic granular sludge under a high organic loading rate [J]. Bioresource Technology Reports, 2019, 7: 100280. doi: 10.1016/j.biteb.2019.100280 [36] 杨华, 黄钧, 赵永贵, 等. 陶厄氏菌Thauera sp. TN9的鉴定及特性 [J]. 应用与环境生物学报, 2013, 19(2): 318-323. doi: 10.3724/SP.J.1145.2013.00318 YANG H, HUANG J, ZHAO Y G, et al. Identif ication and characterization of Thauera sp. strain TN9 [J]. Chinese Journal of Applied and Environmental Biology, 2013, 19(2): 318-323(in Chinese). doi: 10.3724/SP.J.1145.2013.00318
[37] ETCHEBEHERE C, TIEDJE J. Presence of two different active nirS nitrite reductase genes in a denitrifying Thauera sp. from a high-nitrate-removal-rate reactor [J]. Applied and Environmental Microbiology, 2005, 71(9): 5642-5645. doi: 10.1128/AEM.71.9.5642-5645.2005 [38] SRINANDAN C S, SHAH M, PATEL B, et al. Assessment of denitrifying bacterial composition in activated sludge [J]. Bioresource Technology, 2011, 102(20): 9481-9489. doi: 10.1016/j.biortech.2011.07.094 [39] DU S, YA T, ZHANG M L, et al. Distinct microbial communities and their networks in an anammox coupled with sulfur autotrophic/mixotrophic denitrification system [J]. Environmental Pollution, 2020, 262: 114190. doi: 10.1016/j.envpol.2020.114190 -






 DownLoad:
DownLoad: