-
环境内分泌干扰物(EDCs)[1]是20世纪90年代以来引起广泛关注的一类新型化学物质.分为天然和人工合成两类,除铅、汞、砷等金属类外,其余均为有机化学物.按其用途可分为工业原材料、洗涤用品、化妆品、农药、塑料包装和儿童玩具等,因工业生产和使用被不断释放到空气、水体、土壤等环境介质中,可经呼吸空气、饮水、食物摄入及皮肤接触等途径进入人体,影响生物体内激素的合成、释放和代谢,甚至可与激素竞争结合相应受体,或者影响激素受体上下游的调控基因,干扰激素正常的调节功能,造成内分泌系统的紊乱,从而引发各种疾病,对人类健康产生潜在的危害和不利影响[2].
已有大量研究报道EDCs可产生类雌激素效应[3],如邻苯二甲酸脂类、多氯联苯类、双酚类等化合物.这些EDCs在环境浓度下发挥着与天然雌激素类似的生物学活性,从而影响女性的生殖健康[4].已知的不利健康危害包括:月经周期变化、子宫内膜异位症、子宫肌瘤、多囊卵巢综合征、不孕不育、乳腺癌、子宫内膜癌及卵巢癌.此外,近年来的流行病学调查 报告进一步证实女性雌激素敏感癌症发病率与环境中EDCs的浓度存在密切相关性.
EDCs诱导产生的类雌激素效应主要通过经典的核雌激素受体ERα与ERβ介导[5].随着结构生物学研究的深入,有报道证实存在膜雌激素受体(GPER),属于7-跨膜G蛋白偶联受体(GPCR)家族[6]的一员,作为雌激素信号的靶蛋白,参与了雌激素在生殖、神经、内分泌、免疫和心血管系统中的介导作用,对于包括癌症在内的一系列疾病,GPER正成为一种新的治疗靶点和预后指标[7-9].G蛋白偶联雌激素受体(GPER)最初被称为GPR30[10],其结构未被结晶实验确认,激动和拮抗的分子机制及其结构特征的研究仍处在初步阶段.不同于经典的核雌激素受体亚型,GPER被认为是介导快速细胞信号的媒介[11],参与介导了雌激素快速非基因组效应[12].内源性化合物,如雌激素可与细胞膜上的GPER结合,激活快速细胞效应反应[13],包括环磷酸腺苷的产生、细胞内钙离子的动员、多种激酶的激活,如细胞外信号调节激酶、肌醇磷脂3激酶、离子通道以及内皮型一氧化氮合酶等途径.影响下游效应分子在相应的靶组织中发挥其生物学效应[14],从而导致细胞的增值与分化.类似地,环境污染物也可模拟内源性物质结合GPER[15].如BPA可在低浓度下激活GPER,产生较强的雌激素效应[16].
因此,构建快速筛查环境污染物是否结合GPER的分类预测模型,可为理论评估化合物健康风险与毒性效用提供重要的依据.虽然针对环境污染物的毒性预测,已有报道显示机器学习算法可表现出良好的分类预测性能 [17-20],但是结合GPER的小分子却未有可用的分子数据库和已知的预测模型,这限制了构建针对GPER分类预测模型的发展.为了解决这一问题,本研究系统地总结了已报道的有机小分子结合GPER的数据,并进一步评测了随机森林(random forest,RF)、支持向量机(support vector machine,SVM)、神经网络(neural network,ANN)、K-最近邻(K-nearest neighbour,KNN)、朴素贝叶斯(naive bayes,NB)、逻辑回归(logistic,LG)等6种典型机器学习算法的分类预测性能,其中基于RF算法构建的分类模型展示出了优秀的分类预测表现.
-
从文献中查询了130种对GPER检测结合能力的化合物[6, 11, 16, 21-38],分别包括7种双酚类化合物、30种多溴联苯类化合物、18种类固醇类化合物、15种植物激素、30种吲哚及其衍生物、10种农药杀虫剂以及20种GPER选择性配体化合物.其中具有结合能力的化合物有91种;不具有结合能力的化合物有39种.将所有化合物以4∶1的比例随机分成训练集和测试集,其中训练集具有结合能力的有73种,不具有结合能力的有31种;测试集具有结合能力的有18种,不具有结合能力的有8种.利用训练集构建分类预测模型,通过测试集定性评估模型的预测能力.
-
利用软件ChemDraw程序包绘制测试化合物相应的2D分子结构,随后通过Chem 3D程序包转化为相应的3D分子结构,并对其进行能量最小化优化处理.基于优化后的SDF格式的分子结构文件,通过 PaDEL软件计算得到1538个1D和2D分子描述符.根据以下两个原则进行初步的描述符筛选:(1)剔除常数和至少有一个缺失值的描述符;(2)剔除描述符之间相关系数大于0.9的描述符,最终得到369个分子描述符特征.随后利用RF算法和递归特征删减法的描述符重要性评价,将重要性前五位的描述符保留,作为构建机器学习分类预测模型的特征值.
-
为了系统地比较不同机器学习算法的表现,筛选最优算法构建GPER配体分类预测模型,我们选择了6种机器学习算法进行模型的构建[17],分别为:(1)RF算法,是一种集成分类器,它将大量的决策树拟合到一个数据集中,然后将所有树的预测组合起来;(2)SVM算法,通过核函数将输入向量映射到高维特征空间,优化特征空间内的线性划分,构造出具有最大边距的超平面来分离不同类别的数据;(3)ANN算法,采用最常见的前馈反向传播算法,其主要思想是:输入学习样本,使用反向传播算法对网络的权值和偏差进行反复的调整训练,使输出的向量与期望向量尽可能地接近;(4)KNN算法,考虑到数据中的所有情况,并根据局部邻域来获得分类预测.局部领域的大小由参数k决定,即k个最近邻数据点,而类别则由K-最近邻邻居的多数票决定;(5)NB算法,是基于现有的特征并假设特征独立,判断分类类别的概率,进行分类;(6)LG算法,主要研究二分类的响应变量与相应特征值之间的相互关系,并建立相应的预测模型.
所有的数据处理和模型建立都是基于统计编程环境R(版本4.0.2)实现.分别使用randomForest和caret程序包执行特征选择.使用randomForest、kernlab、nnet、kknn、e1071程序包执行RF、SVM、ANN、KNN、NB和LG算法的分类模型构建.
对于分类模型的评估[17],将模型输出的预测分类与实际结果相比,利用混淆矩阵产生真阳性(true positive,TP)、真阴性(true negative,TN)、假阳性(false positive,FP)、假阴性(false negative,FN),并计算相应指标敏感性(Sensitivity,Sn)、特异性(Specificity,Sp)、精确度(Accuracy,Q)以及马修斯相关系数(matthews correlation coefficient,MCC),相应计算公式如下:
敏感性S、特异性Sp、精确度Q以及马修斯相关系数MCC的值越高模型评价越好,精确度在0.9以上为预测结果较好.
而受试者工作特征曲线(receiver operator characteristic curve,ROC)则是另外一个评价二分类模型的重要指标.使用pROC程序包生成ROC曲线并计算每个分类模型的训练集和预测集的AUC值.此外,我们也采用10折交叉验证,来评估模型的精度.最终采用AMBIT Discover软件计算训练集和测试集中化合物的欧几里德距离,评价基于分子描述符的预测模型的应用域.
-
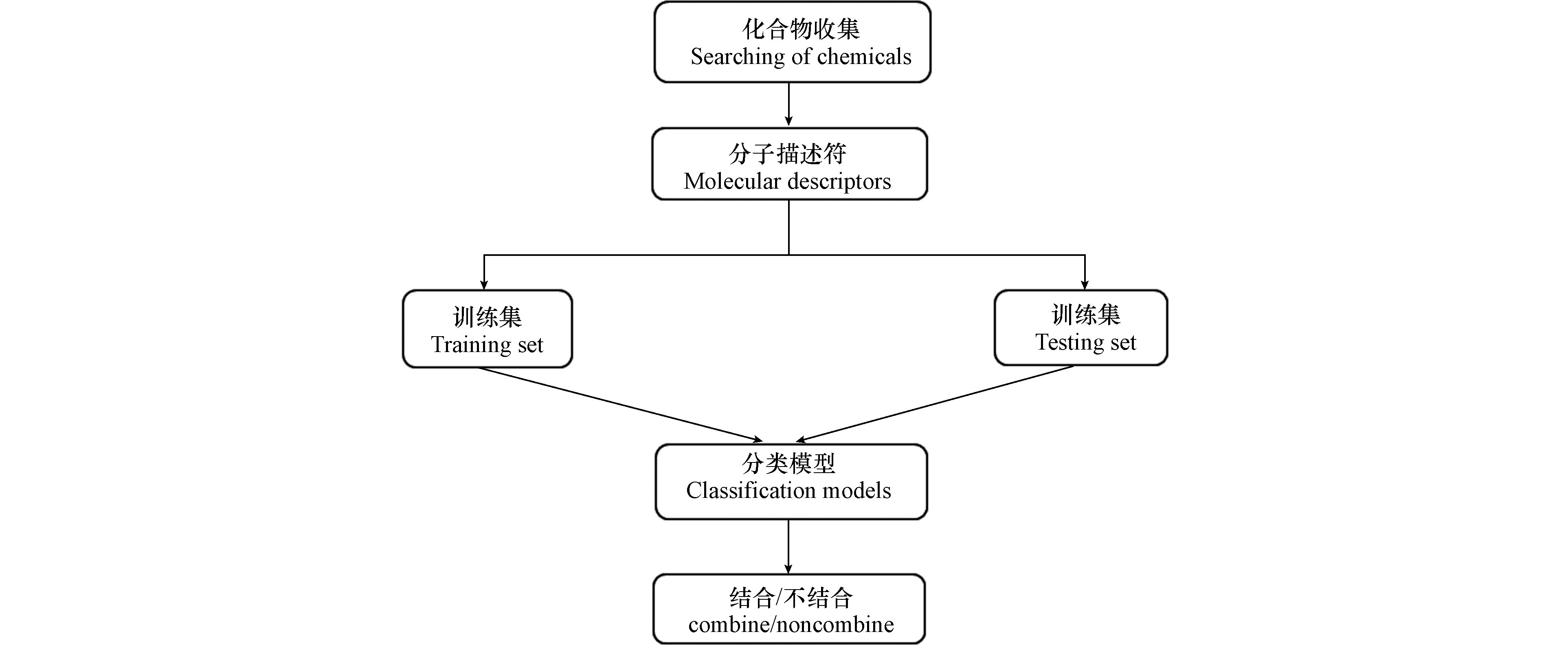
采用10折交叉验证来评估模型的可靠性和预测性能,图1显示了整个分类模型构建的流程.
-
联合RF算法的重要性评价和caret程序包的递归特征删减法进行描述符选取,得到如表1所示的5个最重要的描述符作为模型特征值.
其中,自变量GATS4c、GATS4s、AATSC6i和MATS2i为基于 Geary自相关指数、Broto-Moreau自相关指数以及Moran自相关指数的描述符.自相关指数反应原子性质沿拓扑结构的分布,其中GATS4c和 GATS4s描述符反映了化合物的静电性质对于结合的影响.AATSC6i和MATS2i描述符代表了化合物第一电离能的影响,ETA_Eta表示扩展拓扑化学原子指数对于分子结合能力的影响.
-
对于不同机器学习算法,采用caret程序包的格点搜索法进行超参数的优化,提高分类模型的统计学评价标准.
对于RF模型,通过调整参数trees和mtry的值,改变森林“树”的数量,从而达到对模型的优化效果.trees表示森林“树”的数量,mtry表示每次迭代变量抽样的数量.最终选定RF模型的参数为ntree=500、mtry=5,此时模型预测精度最优.
对于SVM模型,采用高斯RBF核函数进行训练.参数gamma决定了数据映射到新的特征空间后的分布;而参数cost表示为惩罚因子,即对误差的容忍度.最终选定SVM模型的参数为gamma=0.35、cost=10.
针对人工神经网络,分别对size、decay和maxit 等3个参数进行优化,其中size代表隐藏层神经元数,decay代表输入权重的修正参数、maxit代表最大迭代次数.最终选择ANN模型的参数为size=17、decay=0.01、maxit=1000.
对于K-最近邻,通过对邻居数量k的优化,从而增加模型的精度.最终选择5个邻居数的KNN算法建立模型,精确度较高.
对于NB模型,为避免由于样本数据较少而导致整个数据零概率问题的出现,使用拉普拉斯技术做平滑处理,避免零概率问题.
对于LG模型,我们使用逐步回归的方法进行变量重要性的筛选,得到最优模型.
-
通过对模型参数的优化,最终得到最优模型预测结果,如表2所示.
根据结果,表3列举了模型RF、SVM、ANN和KNN的4组预测结果较好的数据,其中Y表示分子结合GPER、N表示不结合.可以看到,测试集的α-E2、训练集的2'-OH-BDE-003和BPF被四个模型全部预测错误.推测原因为α-E2和BPF在结构上与具有结合效应的分子E2和BPA有相似之处,2'-OH-BDE-003在结构上与不具有结合效应的分子BDE-003有相似之处,由此造成了本研究中的所有分类模型都不能对其进行正确的分类.
依据表4,计算模型评价指标Sn、Sp、Q和MCC值,如下所示.可以看到RF的精确度最高,训练集达到99%,测试集达到88.5%,敏感性和特异性也较高,说明RF的预测效果优秀.SVM、ANN、KNN的精度次之,训练集精确度分别达到97.1%、95.2%、95.2%;测试集精确度分别达到88.5%、84.6%、84.6%,敏感性和特异性也较高.而NB和LG的预测结果较差,训练集精度分别为81.7%和79.8%;测试集精度分别为84.6%和88.5%,由此得到最优模型为RF.同时其它模型的统计学评价指标(Sn、Sp、Q和MCC)也显示为RF为最优的分类预测模型.
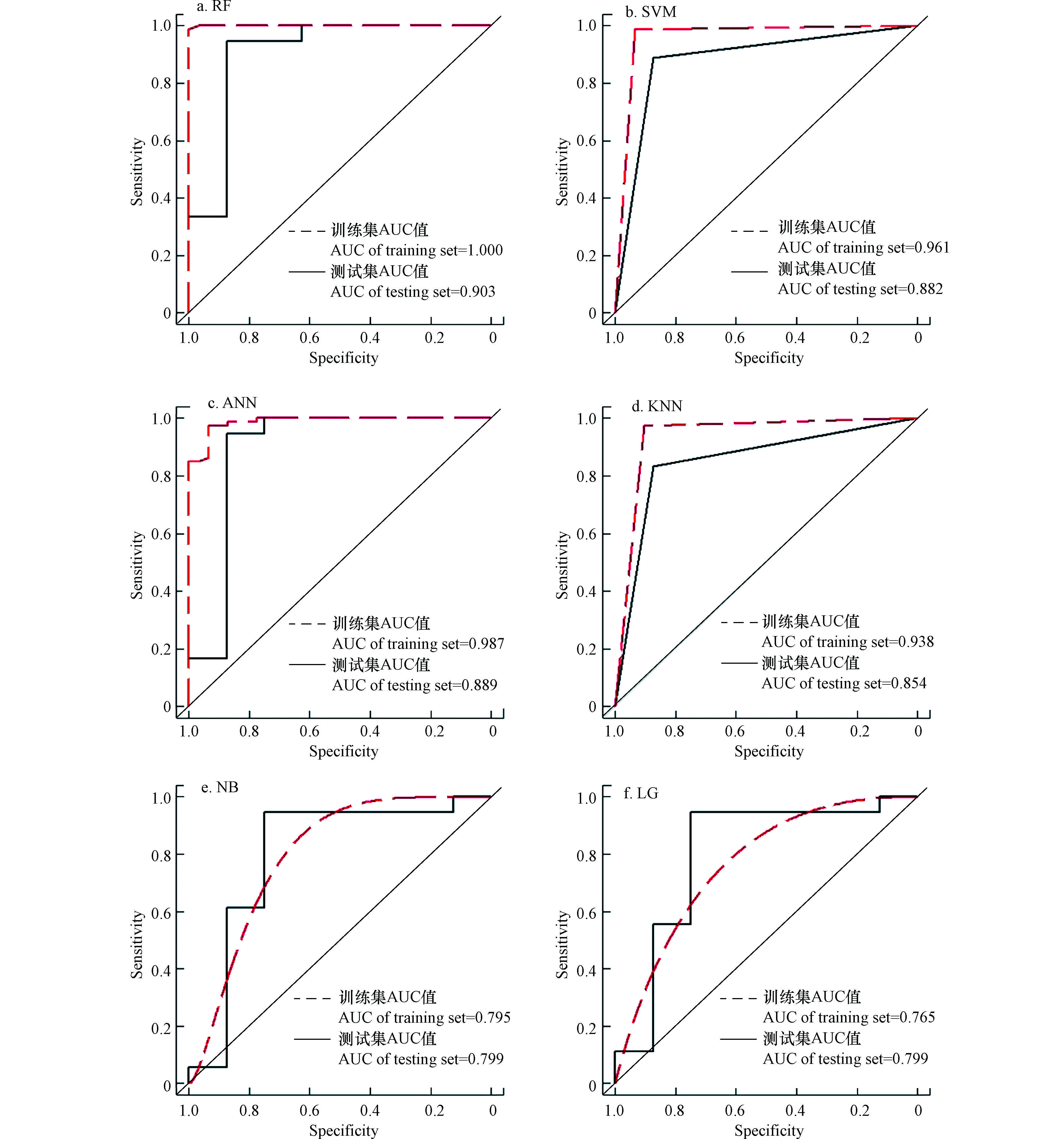
此外通过受试者工作特征(ROC)曲线以及AUC曲下面积对模型评价进行进一步说明.AUC曲下面积越接近1模型性能越高.由图2可知,RF训练集和测试集的AUC值均在0.9以上,说明模型性能最优;SVM、ANN、KNN次之,训练集AUC值分别达到0.961、0.987、0.938,测试集AUC分别达到0.882、0.889、0.854,模型性能较好;NB和LG的训练集AUC值分别为0.795和0.765,测试集AUC均为0.799,说明模型预测性能较差.
最后,通过10折交叉验证来计算模型的精度,其中RF、SVM、ANN、KNN精度分别为81%、81%、81%、80%,而NB和LG的精度较低,分别为75%和72%,说明RF和SVM模型的精度较高,这和前面模型性能评估得出的结论是一致的.
-
本文使用基于欧几里得距离的标准来探索化合物是否适用于所建模型,利用训练集计算可知化合物中最大的欧几里得距离为0.88(截断值),而计算得到的测试集中所有化合物的欧几里得距离均小于0.88,表明测试集化合物均适用于预测模型.这也为测试活性未知的化合物是否适用于所建模型提供了评价标准.
-
目前对于外源性化合物对膜雌激素受体GPER的干扰效应研究方兴未艾,仍有大量潜在的化合物有待验证.因此,在我们的研究中,搜集了130种已知的化合物结合GPER的情况,评价了RF、SVM、ANN、KNN、NB、LG的6种机器学习算法构建的分类预测模型的表现.通过特征选择、模型优化以及模型评价,结果显示RF为最优模型,为评价环境污染物是否通过结合GPER而产生内分泌干扰效应提供了快速筛查的预测模型与理论支撑.
基于膜雌激素受体(GPER)结合化合物能力的分类预测模型
Classification prediction model based on GPER binding ability of membrane estrogen receptor
-
摘要: 近年来,计算毒理学的方法被广泛应用于潜在的环境内分泌干扰物(EDCs)的筛选.膜雌激素受体(GPER),作为一种可以快速响应内源性配体雌激素的关键靶蛋白,调控其介导的多项生理学功能.但是针对GPER的化合物毒性预测模型仍未见报道.因此,本研究收集了130个化合物对GPER的结合活性数据,主要包括双酚类、多溴联苯类以及农药杀虫剂类环境污染物.利用随机森林(RF)、支持向量机(SVM)、人工神经网络(ANN)、K最近邻(KNN)、朴素贝叶斯(NB)以及逻辑回归(LG)等6种机器学习算法构建二分类模型.结果显示,所有被测试算法的测试集准确率均达到85%以上,其中SVM、RF、ANN、KNN等4种算法的训练集准确率高于93%,10折交叉验证准确率高于80%,说明得到的模型具有优秀的分类预测性能.因此,本研究基于机器学习算法构建的分类模型,可以用来快速、准确地预测环境污染物是否通过结合GPER产生内分泌干扰效应.为评估环境污染物的潜在健康风险提供了理论依据.Abstract: In recent years, computational toxicology has been widely applied to the screening of potential environmental endocrine disruptors(EDCs).Membrane estrogen receptor(GPER), as a key target protein that can rapidly respond to endogenous ligand estrogen, regulates its mediated multiple physiological functions. However, the prediction model of compound toxicity for GPER has not been reported. Therefore, the binding activity data of 130 compounds against GPER were collected in this study, mainly including bisphenols, polybrominated biphenyls and environmental pollutants like pesticides and insecticides.Six machine learning algorithms, including random forest (RF), support vector machine(SVM), artificial neural network(ANN), K-nearest neighbor(KNN), Naive Bayes(NB)and logistic regression(LG), were used to construct the dichotomous model.The results showed that the test set accuracy of all the algorithms tested reached more than 85%, among which the training set accuracy of SVM、RF、ANN and KNN algorithm was 93% higher than that of the four algorithms, and the 10-fold cross validation accuracy was 80% higher than that of the four algorithms, indicating that the model obtained had excellent classification prediction performance. Therefore, the classification model built based on machine learning algorithm in this study can be used to quickly and accurately predict whether environmental pollutants will produce endocrine disrupting effects by combining with GPER.It provides a theoretical basis for evaluating the potential health risks of environmental pollutants.
-
沙河水库位于北京市昌平区,上接南沙河、北沙河、东沙河,下连温榆河,是北运河上游干流的关键节点[1]。近年来,周边的生产生活使沙河库区水体污染严重,水库水质仅为《我国地表水环境质量标准》(GB 3838-2002)劣Ⅴ类水体。库区水生植物群落遭到严重破坏,水体自净能力减弱,水生态系统逐渐恶化[2]。因此,亟需开展沙河水库水质提升和水生态修复工作。
水生植物是水生态系统的重要组成部分,参与调控系统的物质循环和能量传递[3],对水生态修复起到关键作用。其中,沉水植物是水生植物群落的重要组成部分,对水体和底泥中的氮、磷等污染物的去除效果良好,同时还能承载水体中的有益微生物[4]。因此,恢复沉水植物群落是水生态系统恢复的关键环节。已有研究表明,沉水植物群落的恢复受温度、pH、营养盐浓度、藻类、风浪等诸多因素影响,其中水体透明度是最关键因素之一[5]。若水体透明度很低,水下光场无法满足沉水植物的生长,植物光合作用受阻而不能成活[6]。现阶段沙河水库水体透明度较低,仅为30~40 cm。因此,提升水体透明度是恢复沙河水库沉水植物群落、修复水库生态系统的先决条件。
投加化学药剂可快速提高水体透明度,但对库区水体内的鱼虾贝壳等水生动物的生长存在风险[7]。近年来,通过投放枝角类等浮游动物的生物操纵手段受到普遍关注[8]。常见的枝角类浮游动物,如大型溞、长肢秀体溞、多刺裸腹溞等,会直接摄食藻类。其分泌物还会促进悬浮颗粒态污染物的沉降,可在短期内提高水体透明度,进而为沉水植物的恢复提供条件[9]。其中,大型溞的应用最为广泛。任文彬[10]在武汉市东湖植物园的研究发现,大型溞在短期内可使湖内水体透明度由50 cm提高至138 cm,效果明显;霍元子等[11]的研究结果表明,大型溞在5 d内便可将上海滴水湖的水体透明度提高至150 cm。上述2个研究中,利用大型溞提高水体透明度的特性,可成功为沉水植物的重建创造条件,从而间接净化水质。而在实际工程应用中,也不乏大型溞提高污染水体透明度的成功案例。如管卫兵等[12]在太湖水域围隔修复区内投放大型溞,利用其对藻类的控制效应,提高了太湖水域的透明度,为后期移栽沉水植物解决太湖水体富营养化问题创造了良好条件。南京市月牙湖通过投放大型溞,后期建立“大型溞-沉水植物”共生生态系统,促进了湖泊生态环境的修复和平衡,使其水质稳定在地表Ⅳ类[13]。
然而,以往通过大型溞提升湖库水体透明度的实际工程中,尚无对大型溞的投放密度开展研究。本研究基于沙河水库的水质改善情况,对大型溞的投放密度参数进行优化,探讨不同投放密度下,大型溞对藻类的摄食及自身的生长情况,并对投溞后沙河水库水体及底泥主要污染物的变化展开研究,以期为后续沉水植物的恢复奠定基础,并为类似湖库的生态修复提供参考。
1. 材料与方法
1.1 实验原料
本实验所用的大型溞购于湖南怀化,为孤雌生殖的纯种大型溞。实验前将其置于沙河水库原水中进行驯化。驯化5 d后,取存活的大型溞进行研究。实验在5月份开展,实验用水和底泥取自北京沙河水库,其COD、TN、NH3-N、
NO−3 1.2 实验装置
装置置于北京林业大学校园内,实物图见图1。装置是长×宽×高为200 cm×50 cm×200 cm的UPVC水箱,水箱侧面为厚度12 mm的UPVC板,正面为厚度为12 mm的有机玻璃板,便于观察投溞后水体透明度的变化。水箱内部分成4个不相互连通的小水箱。小水箱的长×宽×高分别为50 cm×50 cm×200 cm。水箱底部设置泥槽,泥槽的长×宽×高分别为50 cm×50 cm×20 cm。槽底放置沙河水库实际底泥。
1.3 实验方法
将沙河水库底泥混合均匀后铺入各水箱底部泥槽,铺设厚度为5 cm。为保证底泥不受注水过程的剧烈扰动,将取自沙河水库的原水由蠕动泵缓慢抽入4个独立水箱。各水箱注水量为250 L,水深为1 m。2 h后,向水箱内投放不同密度大型溞,投放密度分别为0、5、15和35 ind·L−1。其中,投加密度0为对照组。实验期共35 d,每2 d取各水箱表层水样500 mL,监测主要水质指标有COD、TN、NH3-N、
NO−3 1.4 分析方法
水样COD采用重铬酸钾消解法(GB 11914-1989)、TN采用碱性过硫酸钾消解紫外分光光度法(HJ 636-2012)、NH3-N采用纳氏试剂分光光度法(HJ 535-2009)、
NO−3 底泥中有机质采用重铬酸钾-硫酸消解法(NYT 1121.6-2006)、TN采用凯氏法(HJ 717-2014)、TP采用碱熔-钼锑抗分光光度法(HJ 632-2011)进行测定。
2. 结果与讨论
2.1 不同投放密度下大型溞的繁殖情况
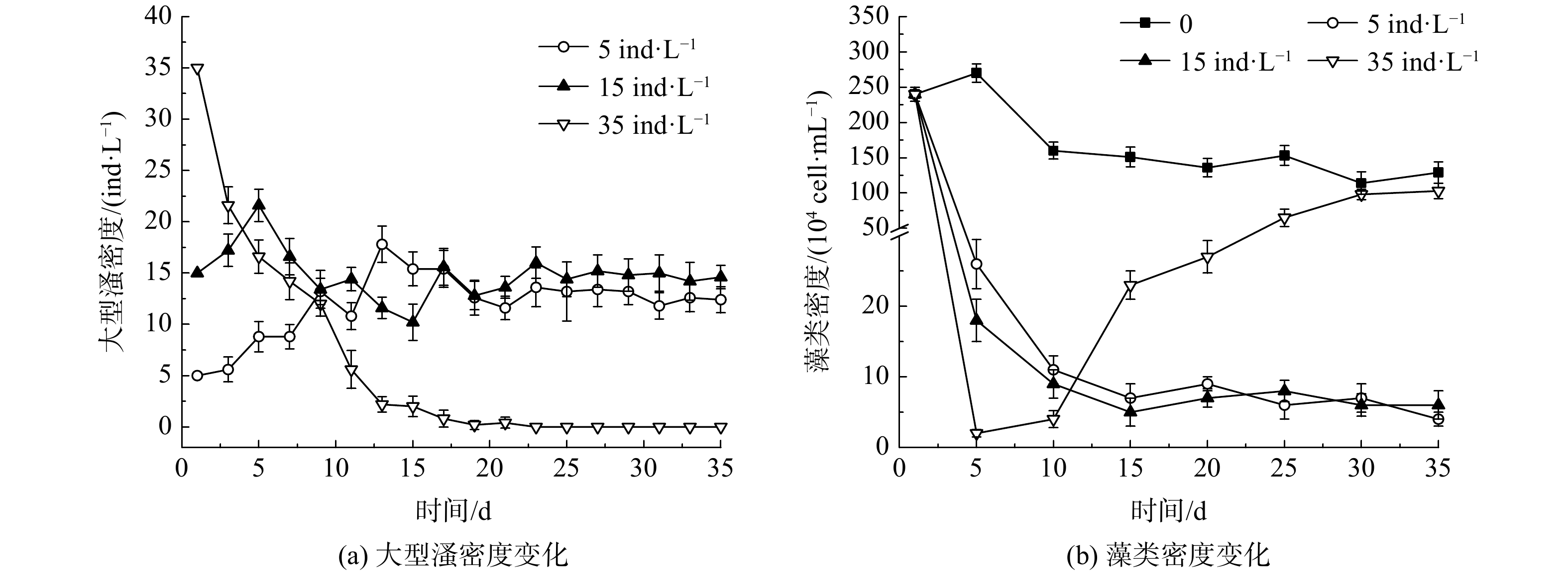
不同投放密度下大型溞的繁殖情况如图2(a)所示。在初始投放密度为5和15 ind·L−1的实验组,大型溞的密度在实验初期上升至18~20 ind·L−1,随后降低,并在实验末期达到相对稳定的状态,密度为14~15 ind·L−1。但当初始投放密度为35 ind·L−1时,一开始水体中大型溞密度会持续下降;投放至10 d时,由起始的35 ind·L−1下降至(5.6±0.3)ind·L−1,15 d后水体中已无法检测到大型溞存在。
藻类是大型溞的主要摄食对象,其数量变化与大型溞的密度变化有密切关系[14]。在不同大型溞投放密度下,水体中藻类的变化如图2(b)所示。投放大型溞后,水体中藻类密度迅速降低,且呈现出大型溞投放密度越大、藻密度下降越快的趋势。当大型溞投放密度为5和15 ind·L−1时,水体中藻密度逐渐下降,并在最终维持在(4~6)×104 cell·mL−1。在此阶段,大型溞生长情况良好,大型溞与藻类之间捕食关系达到动态平衡。而当大型溞投放密度为35 ind·L−1,水体中藻类密度会在第5天骤减至(6±0.2)×104 cell·mL−1,并在5~10 d内维持较低水平;15 d后藻密度逐渐上升,并在实验末期与对照组藻类密度相近。这与大型溞投加密度的变化相关:当大型溞投放密度过大时,短时间将可摄食的藻类捕食殆尽,两者的捕食平衡遭到破坏;之后大型溞由于得不到充足的食物供应而数量急剧下降,甚至死亡;随后,藻类又由于没有了大型溞的捕食,繁殖环境宽松,密度又逐渐上升。
上述结果表明,在适宜的投放密度下,大型溞在沙河水库水样中生长良好,密度维持在14~15 ind·L−1。若初始投放密度过大,大型溞密度会出现不可逆的迅速降低。一方面是由于高密度的大型溞导致种间竞争加剧,大型溞无法得到满足其生长繁殖的食物供应,导致其密度迅速下降[15];另一方面,大型溞种群密度越高,其释放的代谢产物越多,代谢产物的增多会对大型溞的摄食率产生明显的抑制作用,进而影响到大型溞的生长繁殖,促使其失去生命活性产生休眠卵[16]。
2.2 大型溞投放对沙河水库水质的影响
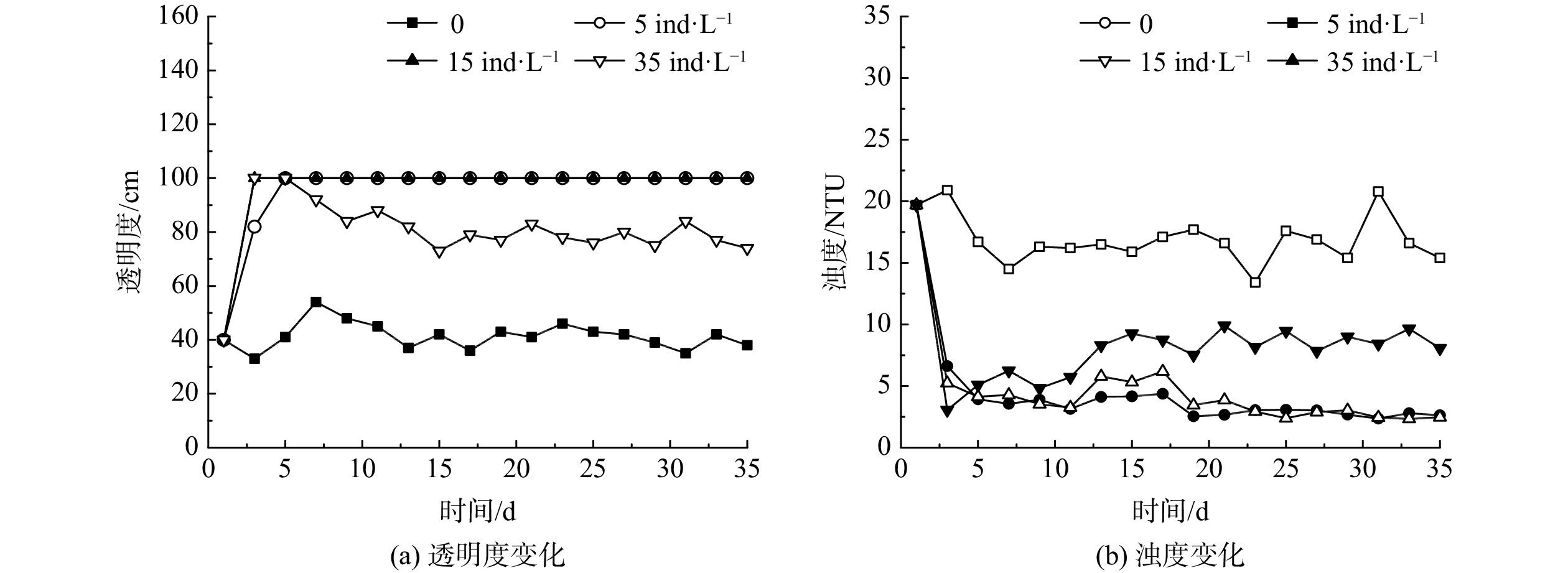
2.2.1 投放密度对水体透明度及浊度的影响
不同大型溞投放密度下水体透明度和浊度的变化如图3所示。对照组水体透明度基本保持不变,为(41.4±4.8)cm,浊度小幅下降后维持在(16.4±1.6)NTU。在大型溞投放密度为5和15 ind·L−1的实验组,水体透明度在投溞后3~5 d内迅速由40 cm升高至100 cm,为对照组的2.5倍;浊度由初始的19.7 NTU下降至(3.4±0.9) NTU。值得注意的是,当投放密度达到35 ind·L−1时,虽然水体透明度在5 d内由40 cm升高至100 cm,浊度也从19.7 NTU下降至5.1 NTU;但在之后的5~15 d,水体透明度由100 cm逐渐下降至73 cm,浊度升高至9.3 NTU;投溞15 d后水体透明度维持在(78.3±3.1) cm,浊度维持在(8.7±0.8) NTU。当初始投放密度达到35 ind·L−1时,由于大型溞在实验后期不断死亡,水体透明度在上升至100 cm后出现了下降;而当投放密度为5和15 ind·L−1时,实验期内大型溞密度稳定,水体透明度可一直维持在100 cm。
上述结果表明,选取适宜的投放密度,投溞可短时间内提高沙河水库水体透明度,降低浊度,并且效果稳定。一方面是由于大型溞会摄食粒径为0.5~40 μm的颗粒物,包括藻类、细菌和碎屑等[17];另一方面,水中悬浮颗粒物可在大型溞排泄物的絮凝作用下成团块状得以沉降,进而从水中得以分离去除[18]。根据实验结果,确定沙河水库大型溞的适宜投放密度为5~15 ind·L−1。
2.2.2 投放密度对水体COD及TP的影响
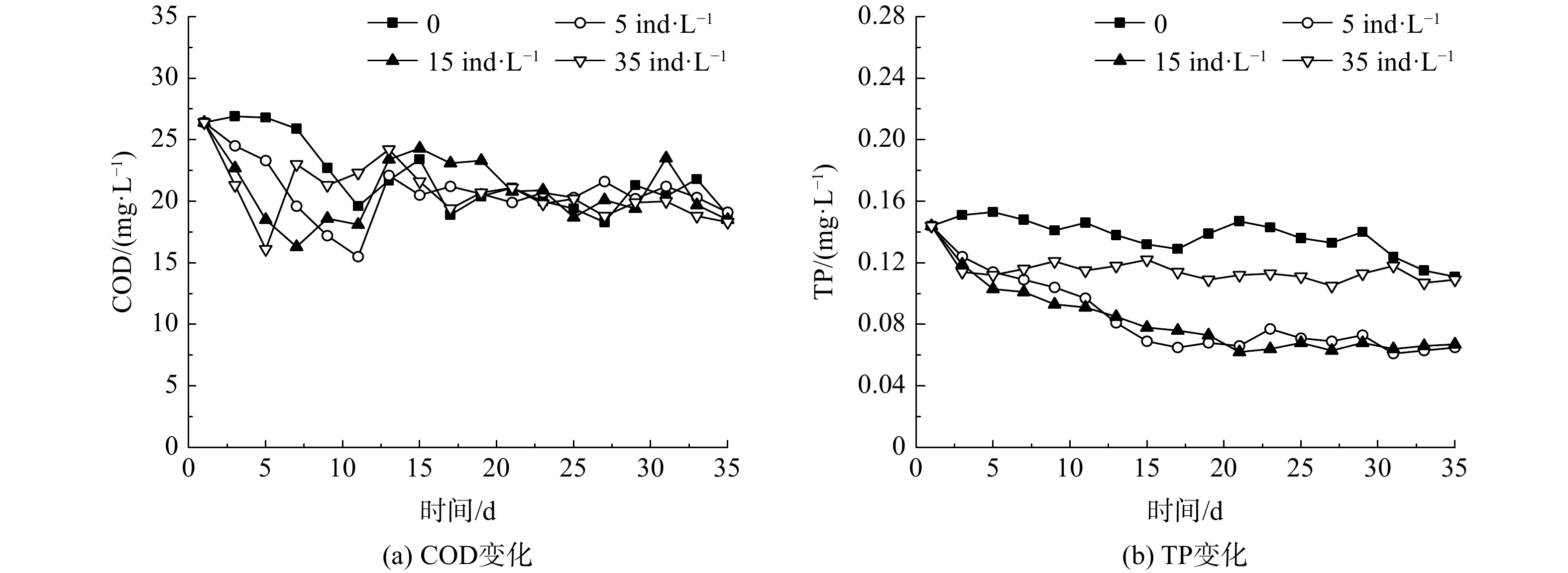
在不同大型溞投放密度下,水体COD和TP的变化如图4所示。由于水体中颗粒态有机物在实验期内发生自然沉降而略有下降,所以对照组的COD也随之下降。各实验组水样的COD在初期均出现了下降,但在5~10 d后,又出现了上升,并在实验后期与对照组相近。COD总体呈现出类似从波动到稳定的趋势,表明大型溞的投放并未引起水体COD的明显变化。
投溞后初期的COD出现下降,是由于大型溞分泌物的絮凝作用使水体中悬浮颗粒态有机物发生沉降。大型溞投放密度为5和15 ind·L−1的水样在之后出现COD上升的现象,可能是因为大型溞在生长过程中的排泄物被微生物分解。当大型溞密度趋于稳定后,COD也相对稳定,只在一定范围内呈现出波动。投放密度为35 ind·L−1的实验组水样中COD出现上升,可能是由于大型溞大量死亡后,其尸体导致水体中COD上升。
在不同的大型溞投放密度下,水体中TP的变化如图4(b)所示。对照组TP较为稳定,保持在(0.137±0.011) mg·L−1。当投放密度为5和15 ind·L−1时,实验过程中TP持续降低,15 d后基本保持稳定;实验结束时,TP由0.144 mg·L−1分别降至0.065 mg·L−1和0.067 mg·L−1,去除率分别为54.9%和53.5%;当投放密度为35 ind·L−1时,TP仅在实验初期1~5 d从0.144 mg·L−1降至0.112 mg·L−1;当大型溞死亡后,水体TP保持稳定,为0.113 mg·L−1。
大型溞不仅可以吸收同化水体中的可溶性磷为自身营养物质[19],其分泌物还可促进水体中悬浮性颗粒态磷的沉降[18]。石岩等[20]在长春南湖投放大型溞,使南湖水体的TP明显下降,去除率达到了89.7%;韩士群等[21]投放长肢秀体溞对水体中TP去除效果明显,去除率为53%。因此,在适宜的投放密度下,大型溞的投放对库区水体TP有较好的去除效果。
2.2.3 投放密度对水体不同形态氮的影响
在不同大型溞投放密度下,3种形态氮素(NH3-N、
NO−3 NO−3 如图5(b)所示,投溞后水体
NO−3 NO−3 NO−3 不同投放密度下,水体TN变化如图5(c)所示。3个实验组中TN从5.79 mg·L−1分别降至1.725、1.883和3.094 mg·L−1,去除率分别达到了70.2%、68.3%和46.5%。投入大型溞会降低水体中TN的浓度已有诸多报道,但不同研究的结果差异较大。张喜勤等[26]研究表明,大型溞对富营养化水体中TN的去除率可高达96.6%;而董旭峰[27]用大型溞净化猪场废水时,TN去除率仅为41.76%。造成差异的原因包括温度、外界干扰条件、水质等多方面的影响。
本研究结果表明,大型溞可通过附着菌群的反硝化作用促进
NO−3 2.3 大型溞投放对库区底泥的影响
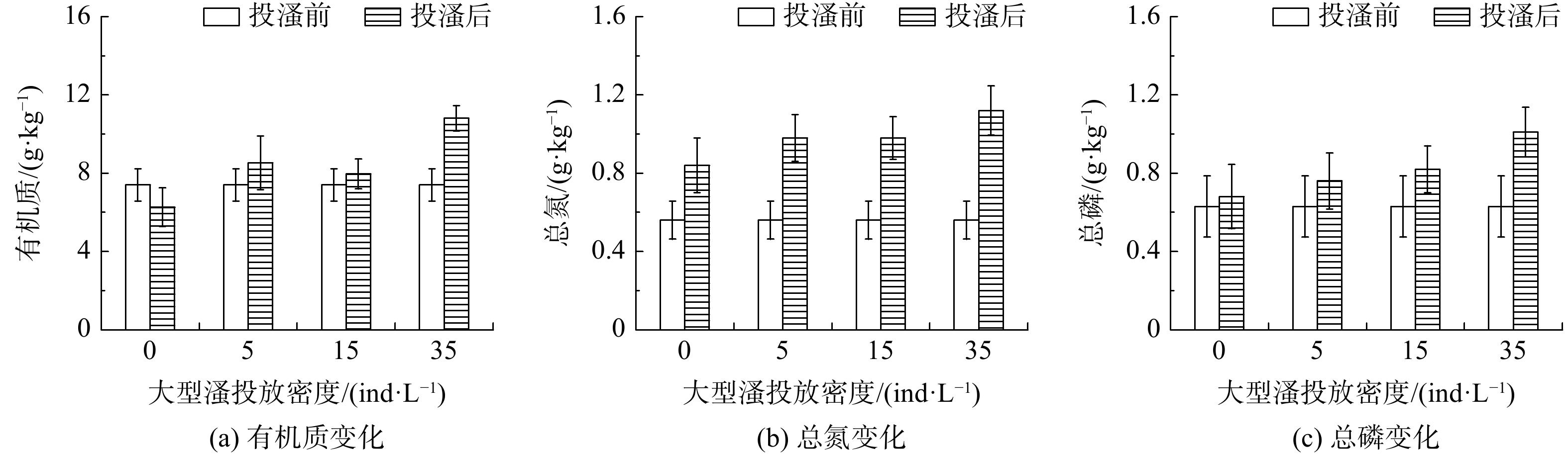
在不同投放密度下,大型溞对沙河水库底泥有机质、TN、TP的影响如图6所示。大型溞投放后,各实验组的有机质均高于对照组,投放密度为5、15和35 ind·L−1的实验组底泥中有机质含量从初始的7.4 g·kg−1分别升高至8.53、7.96和10.81 g·kg−1,分别增加了15.3%、7.7%和46.1%。在不同投放密度下,底泥TN变化如图6(b)所示。对照组总氮由0.56 g·kg−1升高至0.84 g·kg−1,增加了50%,主要是由于水体静止后加速了颗粒态氮的沉降。而投加大型溞的实验组促进了水体中氮素向底泥的沉降作用,实验结束时,底泥总氮的含量分别为0.98、0.98和1.12 g·kg−1,增加率分别为75%、75%和100%。与总氮变化规律一致,底泥中的TP也呈现出不断升高的趋势,不同投放密度实验组的底泥总磷含量分别增加了20.6%、28.5%和60.3%。
由前述分析可知,大型溞分泌的排泄物具有絮凝作用,可使水中的悬浮物沉降,进而引起底泥有机质、TN及TP的增加。投放密度为5和15 ind·L−1的实验组底泥各指标增幅相近,而投放密度为35 ind·L−1的实验组底泥各指标的增幅要远高于前2个实验组。这是由于,在实验中后期大型溞不断死亡,其尸体以及产生的休眠卵发生沉降进入底泥中,故该实验组底泥各指标上幅较大。
3. 结论
1)大型溞在沙河水库水样中可正常生长繁殖。在一定的初始投放密度范围内(5~15 ind·L−1),大型溞保持稳定的生存状态,最终密度为13~14 ind·L−1。经过大型溞的摄食作用,藻类密度最终控制在4×104~6×104 cell·mL−1。
2)投放大型溞利用其摄食藻类、分泌物促进悬浮颗粒态污染物沉降等特性,可在短期内将透明度从40 cm提升至100 cm,并使浊度由最初的19.7 NTU下降至(3.4 ± 0.9) NTU,可为种植水生植物提供良好的先决条件。
3)投溞后,水体中COD并未发生明显改变。由于大型溞促进悬浮颗粒态氮、磷的沉降,TN和TP的去除率分别可达70.2%和54.9%。大型溞投放使实验水体中NH3-N浓度升高32.2%,对水体
NO−3 -
表 1 模型选定的描述符
Table 1. Selected descriptors for models
变量Variables 描述Description GATS4c Geary autocorrelation - lag 4 / weighted by charges GATS4s Geary autocorrelation - lag 4 / weighted by I-state ETA_Eta Composite index Eta AATSC6i Average centered Broto-Moreau autocorrelation - lag 6 / weighted by first ionization potential MATS2i Moran autocorrelation - lag 2 / weighted by first ionization potential 表 2 模型预测结果
Table 2. Model prediction results
数据集Dataset 化合物数Chemical number(n) 真阳性True positive(TP) 真阴性True negative(TN) 假阴性False negative(FN) 假阳性False positive(FP) 模型 RF 训练集 104 73 30 0 1 测试集 26 17 6 1 2 模型 SVM 训练集 104 72 29 1 2 测试集 26 16 7 2 1 模型ANN 训练集 104 72 27 1 4 测试集 26 15 7 3 1 模型 KNN 训练集 104 71 28 2 3 测试集 26 15 7 3 1 模型 NB 训练集 104 69 16 4 15 测试集 26 16 6 2 2 模型 LG 训练集 104 70 13 3 18 测试集 26 16 7 2 1 表 3 模型化合物预测情况
Table 3. Prediction of model compounds
序号No. 化合物Compound 参考文献Reference GPER配体分子(是/否) GPER ligand molecule(Y/N) 观测值Observed SVM RF ANN KNN 1 SK0* [25] Y Y Y Y Y 2 SK0P [25] Y Y Y Y Y 3 G-1 [6, 21] Y Y Y Y Y 4 G-15 [6,21] Y Y Y Y Y 5 G-36 [6,21] Y Y Y Y Y 6 Oleuropein [24] Y Y Y Y Y 7 Hydroxytyrosol [24] Y Y Y Y Y 8 MIBE [24] Y Y Y Y Y 9 4-hydroxytamoxifen [24] Y Y Y Y Y 10 GPER-L1 [24] Y Y Y Y Y 11 GPER-L2 [24] Y Y Y Y Y 12 17β-estradiol [22] Y Y Y Y Y 13 E3 [24] N N N Y N 14 Tamoxifen [22] Y Y Y Y Y 15 Fulvestrant [22] Y Y Y Y Y 16 Epi* [27] Y Y Y Y Y 17 Epi-prop [27] Y Y Y Y Y 18 Epi-4-prop [27] Y Y Y Y Y 19 Epi-5-prop [27] Y Y Y Y Y 20 Epi-Ms [27] N N N Y N 21 C4PY [26] Y Y Y Y Y 22 7β-OH-EpiA* [24] Y Y Y Y Y 23 G-DOTA [37] Y Y Y Y Y 24 G-Bz-DOTA [37] N N N N N 25 G-Bz-DTPA [37] N N N N N 26 Atrazine [24] Y Y Y Y Y 27 PBX1 [34] Y Y Y Y Y 28 PBX2 [34] Y Y Y Y Y 29 ZINC65156419(1) [29] Y Y Y Y N 30 ZINC65156419(2) [29] N N N N N 31 ZINC65156419(3) [29] N N N N N 32 ZINC65156419(4) [29] N N N N N 33 ZINC65156419(5) [29] Y Y Y Y Y 34 ZINC65156419(6) [29] N N N N N 35 ZINC65156419(7) [29] N N N N N 36 ZINC65156419(8) [29] N N N N N 37 ZINC65156419(9)* [29] Y Y Y N Y 38 E2-NH3+ [13] Y Y Y Y Y 39 E2-COO- [13] Y Y Y Y Y 40 E2-NMe3+ [13] Y Y Y Y Y 41 E2-NB [13] Y Y Y Y Y 42 o,p'-DDE [6] Y Y Y Y Y 43 E1* [24] N N Y N N 44 α-E2 [24] N Y Y Y Y 45 Genistein [6] Y Y Y Y Y 46 p,p'-DDT [6] Y Y Y Y Y 47 BPA* [6] Y Y Y Y Y 48 quercetin [24] Y Y Y Y Y 49 Resveratrol* [24] Y Y Y Y Y 50 Raloxifene [24] Y Y Y Y Y 51 zearalonone [6] Y Y Y Y Y 52 Nonylphenol [6] Y Y Y Y Y 53 kepone [6] Y Y Y Y Y 54 STX [24] Y Y Y Y Y 55 PPT* [24] Y Y Y Y Y 56 2,2',5'-PCB-4-OH [6] Y Y Y Y Y 57 equol [24] Y Y Y Y Y 58 2-methoxye stradiol [24] Y Y Y Y Y 59 niacin [24] Y Y Y Y Y 60 daidzein [24] Y Y Y Y Y 61 BDE-003 [28] N N N N N 62 BDE-007* [28] N N N N N 63 BDE-028 [28] N N N N N 64 BDE-047 [28] N N N N N 65 BDE-049* [28] N N N N N 66 BDE-085* [28] N N N N N 67 BDE-099* [28] N N N N N 68 BDE-100 [28] N N N N N 69 BDE-154 [28] N N N N N 70 BDE-180 [28] N N N N N 71 BDE-187 [28] N N N N N 72 BDE-201 [28] N N N N N 73 2'-OH-BDE-003* [28] Y N N N N 74 3'-OH-BDE-007 [28] Y Y Y Y Y 75 3'-OH-BDE-028 [28] Y Y Y Y Y 76 3'-OH-BDE-047* [28] Y Y Y Y Y 77 3'-OH-BDE-154 [28] Y Y Y Y Y 78 4'-OH-BDE-049 [28] Y Y Y Y Y 79 5'-OH-BDE-099* [28] Y Y Y Y Y 80 2'-OH-BDE-007 [28] N N N N N 81 2'-OH-BDE-028* [28] N N N N N 82 3-OH-BDE-100 [28] Y N Y N Y 83 4-OH-BDE-187 [28] Y Y Y Y Y 84 4'-OH-BDE-201 [28] Y Y Y Y Y 85 5-OH-BDE-047 [28] N N N N N 86 5'-OH-BDE-100 [28] N N N N N 87 6-OH-BDE-047 [28] N N N N N 88 6-OH-BDE-085* [28] N N N N N 89 6'-OH-BDE-099 [28] N N N N N 90 6-OH-BDE-180 [28] Y Y Y Y N 91 BPAF [16] Y Y Y Y Y 92 BPB* [16] Y Y Y Y Y 93 BPF* [16] N Y Y Y Y 94 BPS* [16] Y Y Y N N 95 TBBPA [16] N N N N Y 96 TCBPA [16] Y Y Y Y Y 97 Diethylstilbestro [24] N N N N N 98 2-Hydroxy stradiol* [24] Y N Y Y N 99 Aldosterone [24] Y Y Y Y Y 100 Tectoridin [24] Y Y Y Y Y 101 Apigenin* [24] Y Y Y Y Y 102 Methoxychlor [24] Y Y Y Y Y 103 p,p'-DDE* [24] Y Y Y Y Y 104 o,p'-DDT* [24] Y Y Y Y Y 105 DPN [24] Y Y Y Y Y 106 Ethynylestradiol [24] Y Y Y Y Y 107 3MC [32] Y Y Y Y Y 108 AB-1 [31] N N N N N 109 CIMBA-5* [36] Y Y Y Y Y 110 CIMBA-6 [36] Y Y Y Y Y 111 CIMBA-7 [36] Y Y Y Y Y 112 CIMBA-8 [36] Y Y Y Y Y 113 CIMBA-9 [36] N Y N Y N 114 CIMBA-10 [36] Y Y Y Y Y 115 CIMBA-11 [36] Y Y Y Y Y 116 CIMBA-12 [36] Y Y Y Y Y 117 CIMBA-13 [36] Y Y Y Y Y 118 CIMBA-14 [36] N N N N N 119 CIMBA-15 [36] Y Y Y Y Y 120 CIMBA-16 [36] N N N N Y 121 CIMBA-17 [36] Y Y Y Y Y 122 CIMBA-18 [36] Y Y Y Y Y 123 CIMBA-19 [36] N N N N N 124 CIMBA-20 [36] Y Y Y Y Y 125 CIMBA-21 [36] Y Y Y Y Y 126 CIMBA-22 [36] Y Y Y Y Y 127 CIMBA-23 [36] Y Y Y Y Y 128 CIMBA-24 [36] Y Y Y Y Y 129 CIMBA-25 [36] Y Y Y Y Y 130 Carbhydraz [35] Y Y Y Y Y 注:*测试集化合物. Note:*Testing set compounds. 表 4 模型评价
Table 4. Model performance
数据集Dataset 化合物数Chemical number(n) 敏感性Sensitivity(Sn) 特异性Specificity(Sp) 精确度Accuracy(Q) 马修斯相关系数Matthews correlation coefficient(MCC) 模型 RF 训练集 104 1 0.968 0.99 0.977 测试集 26 0.944 0.75 0.885 0.723 模型 SVM 训练集 104 0.986 0.935 0.971 0.931 测试集 26 0.889 0.875 0.885 0.741 模型ANN 训练集 104 0.986 0.871 0.952 0.884 测试集 26 0.833 0.875 0.846 0.672 模型 KNN 训练集 104 0.972 0.903 0.952 0.884 测试集 26 0.833 0.875 0.846 0.672 模型 NB 训练集 104 0.945 0.516 0.817 0.535 测试集 26 0.889 0.75 0.846 0.639 模型 LG 训练集 104 0.959 0.419 0.798 0.480 测试集 26 0.889 0.875 0.885 0.741 -
[1] FENICHEL P, CHEVALIER N. Is testicular germ cell cancer estrogen dependent? The role of endocrine disrupting chemicals [J]. Endocrinology, 2019, 160(12): 2981-2989. doi: 10.1210/en.2019-00486 [2] BALABANIC D, RUPNIK M S, KLEMENCIC A K. Negative impact of endocrine-disrupting compounds on human reproductive health [J]. Reproduction Fertility and Development, 2011, 23(3): 403-416. doi: 10.1071/RD09300 [3] BRAUN J M. Early-life exposure to EDCs: Role in childhood obesity and neurodevelopment [J]. Nature Reviews Endocrinology, 2017, 13(3): 161-173. doi: 10.1038/nrendo.2016.186 [4] FOWLER P A, BELLINGHAM M, SINCLAIR K D, et al. Impact of endocrine-disrupting compounds (EDCs) on female reproductive health [J]. Molecular and Cellular Endocrinology, 2012, 355(2): 231-239. doi: 10.1016/j.mce.2011.10.021 [5] HARRIS H A, KATZENELLENBOGEN J A, KATZENELLENBOGEN B S J E. Characterization of the biological roles of the estrogen receptors, ERα and ERβ, in estrogen target tissues in vivo through the use of an ERα-selective ligand [J]. Endocrinology, 2002(11): 4172-4177. [6] THOMAS P, DONG J. Binding and activation of the seven-transmembrane estrogen receptor GPR30 by environmental estrogens: A potential novel mechanism of endocrine disruption [J]. The Journal of Steroid Biochemistry and Molecular Biology, 2006, 102(1): 175-179. [7] PROSSNITZ E R, BARTON M. The G-protein-coupled estrogen receptor GPER in health and disease [J]. Nature Reviews Endocrinology, 2011, 7(12): 715-726. doi: 10.1038/nrendo.2011.122 [8] PROSSNITZ E R, BARTON M. Signaling, physiological functions and clinical relevance of the G protein-coupled estrogen receptor GPER [J]. Prostaglandins Other Lipid Mediat, 2009, 89(3-4): 89-97. doi: 10.1016/j.prostaglandins.2009.05.001 [9] WANG C, LI J, YE S, et al. Oestrogen inhibits VEGF expression and angiogenesis in triple-negative breast cancer by activating GPER-1 [J]. Journal of Cancer, 2018, 9(20): 3802-3811. doi: 10.7150/jca.29233 [10] NIRO S, PEREIRA E, PELISSIER M A, et al. The DHEA metabolite 7β-hydroxy-epiandrosterone exerts anti-estrogenic effects on breast cancer cell lines [J]. Steroids, 2012, 77(5): 542-551. doi: 10.1016/j.steroids.2012.01.019 [11] PROSSNITZ E, BARTON M. Estrogen biology: New insights into GPER function and clinical opportunities [J]. Molecular and Cellular Endocrinology, 2014, 389(1): 71-83. [12] ALBANITO L, MADEO A, LAPPANO R, et al. G protein-coupled receptor 30 (GPR30) mediates gene expression changes and growth response to 17β-estradiol and selective GPR30 ligand G-1 in ovarian cancer cells [J]. Cancer Research, 2007, 67(4): 1859-1866. doi: 10.1158/0008-5472.CAN-06-2909 [13] REVANKAR C M, MITCHELL H D, FIELD A S, et al. Synthetic estrogen derivatives demonstrate the functionality of intracellular GPR30 [J]. Acs Chemical Biology, 2007, 2(8): 536-544. doi: 10.1021/cb700072n [14] TSUGAWA Y, HIRAMOTO M. IMAI T. Estrogen induces estrogen receptor α expression and hepatocyte proliferation in late pregnancy [J]. Biochemical & Biophysical Research Communications, 2019, 511(3): 592-596. [15] PROSSNITZ E R. GPER modulators: Opportunity nox on the heels of a class akt [J]. The Journal of Steroid Biochemistry and Molecular Biology, 2018, 176: 73-81. doi: 10.1016/j.jsbmb.2017.03.005 [16] CAO L Y, REN X M, LI C H, et al. Bisphenol AF and bisphenol B exert higher estrogenic effects than bisphenol A via G protein-coupled estrogen receptor pathway [J]. Environmental Science & Technology, 2017, 51(19): 11423-11430. [17] RUSSO D P, ZORN K M, CLARK A M, et al. Comparing multiple machine learning algorithms and metrics for estrogen receptor binding prediction [J]. Molecular Pharmaceutics, 2018, 15(10): 4361-4370. doi: 10.1021/acs.molpharmaceut.8b00546 [18] GRISONI F, CONSONNI V. BALLABIO D. Machine learning consensus to predict the binding to the androgen receptor within the compara project [J]. Journal of Chemical Information and Modeling, 2019, 59(5): 1839-1848. doi: 10.1021/acs.jcim.8b00794 [19] SHEFFIELD T, JUDSON R. Ensemble QSAR modeling to predict multispecies fish toxicity lethal concentrations and points of departure [J]. Environmental Science & Technology, 2019, 53(21): 12793-12802. [20] WANG Z, CHEN J, HONG H. Applicability domains enhance application of PPARγ agonist classifiers trained by drug-like compounds to environmental chemicals [J]. Chemical Research in Toxicology, 2020, 33(6): 1382-1388. doi: 10.1021/acs.chemrestox.9b00498 [21] ARNATT C K, ZHANG Y. G protein-coupled estrogen receptor (GPER) agonist dual binding mode analyses toward understanding of its activation mechanism: a comparative homology modeling approach [J]. Molecular Informmatics, 2013, 32(7): 647-658. doi: 10.1002/minf.201200136 [22] WANG D, HU L, ZHANG G, et al. G protein-coupled receptor 30 in tumor development [J]. Endocrine, 2010, 38(1): 29-37. doi: 10.1007/s12020-010-9363-z [23] BARTON M, PROSSNITZ E R. Emerging roles of GPER in diabetes and atherosclerosis [J]. Trends in Endocrinology & Metabolism, 2015, 26(4): 185-192. [24] PROSSNITZ E, ARTERBURN J. International union of basic and clinical pharmacology. XCVII. G protein-coupled estrogen receptor and its pharmacologic modulators [J]. Pharmacological Reviews, 2015, 67(3): 505-540. doi: 10.1124/pr.114.009712 [25] KHAN S U, AHEMAD N, CHUAH L H, et al. Sequential ligand and structure-based virtual screening approach for the identification of potential G protein-coupled estrogen receptor-1 (GPER-1) modulators [J]. Rsc Advances, 2019, 9(5): 2525-2538. doi: 10.1039/C8RA09318K [26] LAPPANO R, ROSANO C, PISANO A, et al. A calixpyrrole derivative acts as an antagonist to GPER, a G-protein coupled receptor: mechanisms and models [J]. Disease models & mechanisms, 2015, 8(10): 1237-1246. [27] SARMIENTO V, SANCHEZ T R, ULLOA A M, et al. Synthesis of novel (-) -epicatechin derivatives as potential endothelial GPER agonists: Evaluation of biological effects [J]. Bioorganic & Medicinal Chemistry Letters, 2018, 28(4): 658-663. [28] CAO L Y, REN X M, YANG Y, et al. Hydroxylated polybrominated biphenyl ethers exert estrogenic effects via non-genomic G protein-coupled estrogen receptor mediated pathways [J]. Environmental Health Perspectives, 2018, 126(5): 057005. doi: 10.1289/EHP2387 [29] O’DEA A, SONDERGARD C, SWEENEY P, et al. A series of indole-thiazole derivatives act as GPER agonists and inhibit breast cancer cell growth [J]. Acs Medicinal Chemistry Letters, 2018, 9(9): 901-906. doi: 10.1021/acsmedchemlett.8b00212 [30] ALBANITO L, LAPPANO R, MADEO A, et al. Effects of atrazine on estrogen receptor α and G protein-coupled receptor 30-mediated signaling and proliferation in cancer cells and cancer-associated fibroblasts [J]. Environmental Health Perspectives, 2015, 123(5): 493-499. doi: 10.1289/ehp.1408586 [31] REVANKAR C M, BOLOGA C G, PEPERMANS R A, et al. A selective ligand for estrogen receptor proteins discriminates rapid and genomic signaling [J]. Cell Chemical Biology, 2019, 26(12): 1692-1702. doi: 10.1016/j.chembiol.2019.10.009 [32] CIRILLO F, LAPPANO R, BRUNO L, et al. AHR and GPER mediate the stimulatory effects induced by 3-methylcholanthrene in breast cancer cells and cancer-associated fibroblasts (CAFs) [J]. Journal of Experimental & Clinical Cancer Research, 2019, 38(1): 335-353. [33] ALDO M U, DAVID M L, ERNESTO B R, et al. The effects of (-)-epicatechin on endothelial cells involve the G protein-coupled estrogen receptor (GPER) [J]. Pharmacological Research, 2015, 100: 309-320. doi: 10.1016/j.phrs.2015.08.014 [34] MAGGIOLINI M, SANTOLLA M, AVINO S, et al. Identification of two benzopyrroloxazines acting as selective GPER antagonists in breast cancer cells and cancer-associated fibroblasts [J]. Future medicinal chemistry, 2015, 7(4): 437-448. doi: 10.4155/fmc.15.3 [35] ROSANO C, PONASSI M, SANTOLLA M F, et al. Macromolecular modelling and docking simulations for the discovery of selective GPER ligands [J]. The AAPS Journal, 2016, 18(1): 41-46. doi: 10.1208/s12248-015-9844-3 [36] DELEON C, WANG H, GUNN J, et al. A novel GPER antagonist protects against the formation of estrogen-induced cholesterol gallstones in female mice [J]. Journal of Lipid Research, 2020, 61(5): 767-777. doi: 10.1194/jlr.RA119000592 [37] NAYAK T K, DENNIS M K, RAMESH C, et al. Influence of charge on cell permeability and tumor imaging of GPR30-targeted 111in-labeled nonsteroidal imaging agents [J]. ACS Chemical Biology, 2010, 5(7): 681-690. doi: 10.1021/cb1000636 [38] LUO L J, LIU F, LIN Z K, et al. Genistein regulates the IL-1 beta induced activation of MAPKs in human periodontal ligament cells through G protein-coupled receptor 30 [J]. Archives of Biochemistry and Biophysics, 2012, 522(1): 9-16. doi: 10.1016/j.abb.2012.04.007 -






 下载:
下载: