-
吲哚常见于焦化废水中,是一种典型的氮杂环污染物。环境中过量的吲哚类物质会对人体造成伤害,故对吲哚废水的处理与资源化已成为急须解决的问题[1-4]。已有研究[5-11]表明,微生物能够对吲哚进行生物转化,生成靛蓝、靛玉红等具有高附加值的靛蓝类色素。相关研究有助于探索吲哚废水的资源化处置,近年来已逐渐引起人们的广泛关注。
研究发现,苯酚降解菌及相关功能酶能够较好地转化吲哚合成靛蓝类色素。KIM等[12]从苯酚降解菌Pseudomonas sp. KL28中克隆表达的多组分苯酚羟化酶(mPHKL28)能够转化吲哚,合成靛蓝和7-羟基吲哚,也能转化多种吲哚衍生物,合成具有不同颜色的靛蓝类色素。QU等[13]分离得到的苯酚降解菌Pseudomonas monteilii QM可转化吲哚,合成靛蓝、靛玉红等物质。WANG等[14]筛选的Pseudomonas sp. PI1和Acinetobacter sp. PI2也可在苯酚诱导下转化吲哚,合成多种靛蓝类色素。目前,苯酚降解菌对吲哚的生物转化研究报道较多,但是对于其他吲哚类物质(如甲基吲哚)的生物转化研究相对较少。
本研究在土壤中分离得到1株苯酚降解菌YC,通过形态观察及16S rRNA序列分析对菌株进行鉴定,考察菌株对吲哚及甲基吲哚的生物转化特性,并利用响应曲面法对菌株转化吲哚合成靛蓝的条件进行优化,为焦化废水中吲哚的生物处理与资源化提供高效的微生物资源。
-
采用无机盐培养基,其组成包括Na2HPO4·12H2O 3.28 mg·L−1、KH2PO4 2 mg·L−1、(NH4)2SO4 2 mg·L−1、FeCl3 0.25 mg·L−1,pH=7.0。
实验所用吲哚、甲基吲哚、靛蓝、二甲基亚砜及无机盐培养基成分均为分析纯,甲醇为色谱纯,土壤样品取自大连理工大学盘锦校区校园。
使用的主要仪器包括:场发射扫描电子显微镜(Nova NanoSEM 450,美国FEI公司);紫外-可见分光光度计(UH5300,日本HITACHI公司);高效液相色谱仪(1290 Infinity II,德国Agilent公司);四级杆轨道离子阱高分辨质谱仪(Q Exactive,美国Thermo Fisher科技有限公司)。
-
取10 g土壤样品加入100 mL含苯酚(100 mg·L−1)的无机盐培养基,好氧培养7 d后,按体积分数10%接种量转接到新的无机盐培养基(含苯酚100 mg·L−1)中继续培养。经3~4次培养后,取驯化后的样品反复进行平板涂布,最后得到具有苯酚降解能力的单菌落,命名为YC。利用扫描电子显微镜观察菌株的细胞形态。
以菌株YC菌落为模板,选择16S rRNA通用引物对其16S rRNA基因序列进行PCR扩增,送至生工生物工程(上海)股份有限公司检测,测序结果经Blast在Genbank中比对分析。利用MEGA 6.0软件构建系统发育树,确定菌株的分类。
采用含3 g·L−1酵母浸粉与100 mg·L−1苯酚的无机盐培养基进行富集培养,好氧培养24 h后,离心收集菌体(10 000 r·min−1,10 min),使用无机盐培养基,洗涤2次后备用。
-
将收集好的菌体用无机盐培养基重悬,充分摇匀至无固形物,采用紫外-可见分光光度计测定菌悬液OD660,并利用无机盐培养基调节OD660为2.0。移取20 mL菌悬液,分别加入50 mg·L−1吲哚或甲基吲哚,好氧培养24 h,取反应液进行后续分析。
-
采用响应曲面法对菌株YC转化吲哚合成靛蓝进行条件优化。响应曲面法可通过建模分析,在最低限度的实验次数下,利用最小二分法对多个受变量影响的因素进行分析,并探究因素间的影响关系[15]。本研究以靛蓝产量为响应目标,探究生物量(因素A)、吲哚浓度(因素B)与反应体系pH(因素C)对生物转化过程的影响。采用Design Expert 8.0软件设计实验并进行分析,生物量OD660值为1.5~2.5,吲哚浓度为50~150 mg·L−1,pH为6~8,标号为15、16、17、18、19和20的实验组为各因素中心点值的实验组合。
-
1) 液相色谱(HPLC)分析。取反应液离心,沉淀加入适量二甲基亚砜,重悬离心,取上清液过0.45 μm有机膜后进行HPLC分析。HPLC条件如下:Hypersil ODS2 C18色谱柱(5 μm,4.6 mm×250 mm),流速为1.0 mL·min−1,进样量为5 μL,流动相为水/甲醇;洗脱时间共计20 min,梯度洗脱时间为0~15 min,甲醇含量由60%升至70%;等度洗脱时间为15~20 min,甲醇含量为60%。
2) 液相-质谱(LC-MS)分析。产物分离过程同上,LC条件:Ultimate XB-C18色谱柱(3 μm, 2.1 mm×150 mm),流速为0.4 mL·min−1,进样量为10 μL,流动相为水/甲醇;洗脱时间共计5 min,梯度洗脱时间为0~2 min,甲醇含量由50%升至95%;等度洗脱时间为2~4 min,甲醇含量为95%;梯度洗脱时间为4~5 min,甲醇含量为50%。MS条件:采用ESI离子源,雾化室温度为320 ℃,以N2为保护气和干燥气,进行负模式扫描。
-
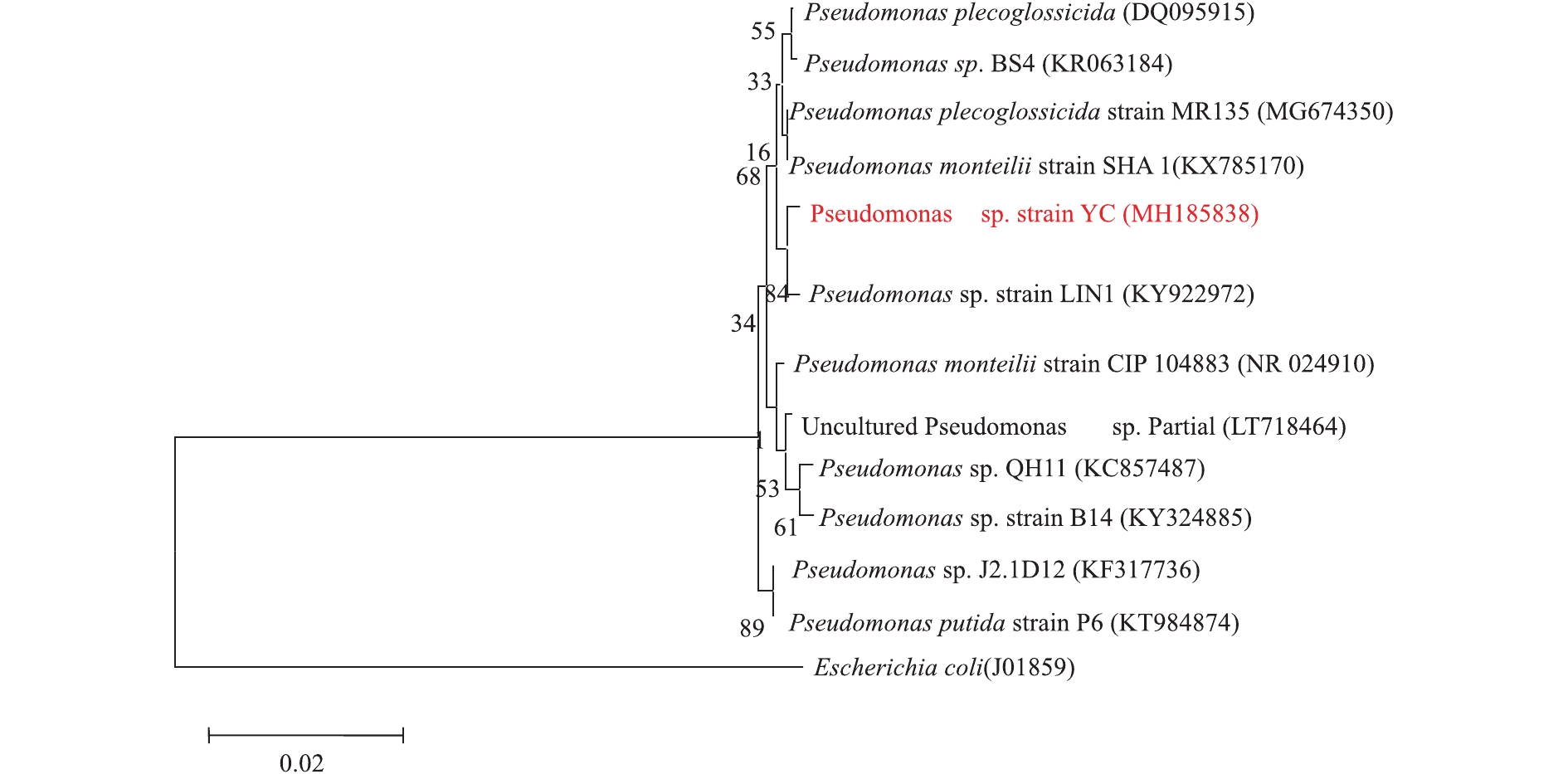
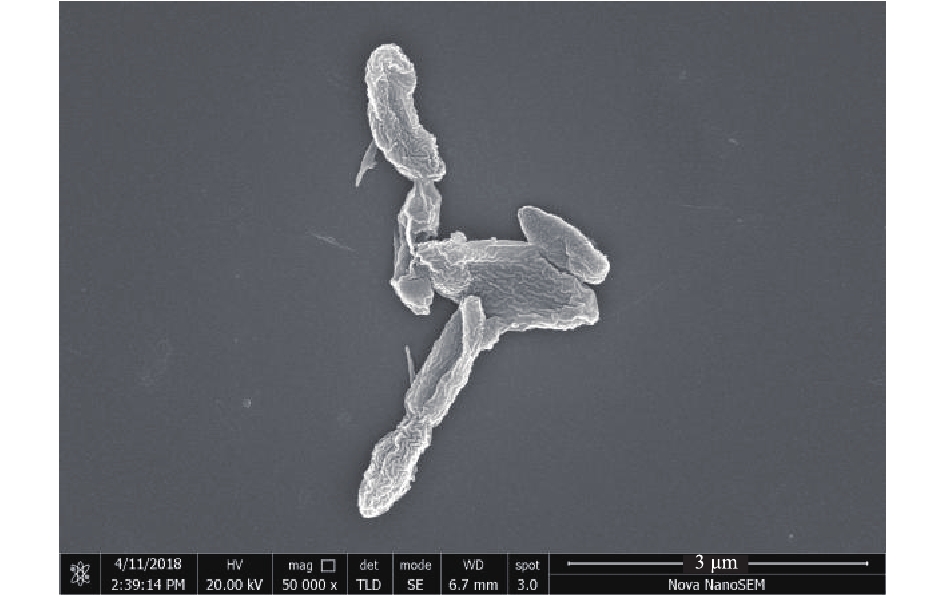
通过反复平板涂布,筛选得到了1株能以苯酚为唯一碳源和能源生长的菌株,命名为YC。菌株YC在固体培养基上的菌落呈白色,为表面光滑且边缘规则的圆形,扫描电镜下为杆状(图1)。通过16S rRNA序列对比发现,菌株YC的16S rRNA(GenBank登录号MH185838)与多株Pseudomonas属菌株的序列相似度为98%以上,因此,菌株YC为Pseudomonas sp.(假单胞菌属)。采用比邻法构建菌株YC的系统发育树(如图2所示),结果显示,菌株YC位于Pseudomonas属簇内,并且与Pseudomonas plecoglossicida菌种及Pseudomonas monteilii菌种的亲缘性更近。
-
实验结果表明,菌株YC在含有苯酚(100 mg·L−1)与吲哚(50 mg·L−1)的无机盐培养基中生长时,能够生成蓝色的气泡,表明可能有靛蓝类色素合成,但菌株生长量较低,靛蓝产量也相对较低。因此,考虑利用添加了酵母浸粉和苯酚的无机盐培养基对菌株进行富集培养,其中酵母浸粉能够为菌株生长提供充足的营养物质,而苯酚能够诱导菌株表达芳烃加氧酶(如苯酚羟化酶)。将培养好的菌株离心收集,并通过无机盐培养基重悬制备休眠细胞,选取吲哚、2-甲基吲哚、3-甲基吲哚、5-甲基吲哚、6-甲基吲哚、7-甲基吲哚,考察菌株YC休眠细胞生物转化吲哚及甲基吲哚合成靛蓝类色素的可行性。由图3可知,菌株YC能够较好地转化吲哚(6号)、5-甲基吲哚(3号)、6-甲基吲哚(4号)、7-甲基吲哚(5号)生产蓝色或紫色的产物,但2-甲基吲哚(1号)的转化产物为黄色,而3-甲基吲哚(2号)的反应后无明显颜色。
吲哚可经氧化生成2-羟吲哚和3-羟吲哚,靛蓝则常由3-羟吲哚二聚形成[3]。相较其他甲基吲哚,2-甲基吲哚由于其2号碳原子位置上的甲基极有可能阻碍苯酚羟化酶在3号碳位置上的加氧羟化,从而阻碍靛蓝类色素的合成,故其并未显著合成靛蓝类色素[12, 16]。而3-甲基吲哚的3号碳位置上的甲基则会完全阻碍了3号碳位置上的羟化,由此推断,其无法通过在3号碳位置上进行羟化得到靛蓝类色素。实验结果表明苯酚降解菌YC具有较为广谱的靛蓝类色素合成能力。
-
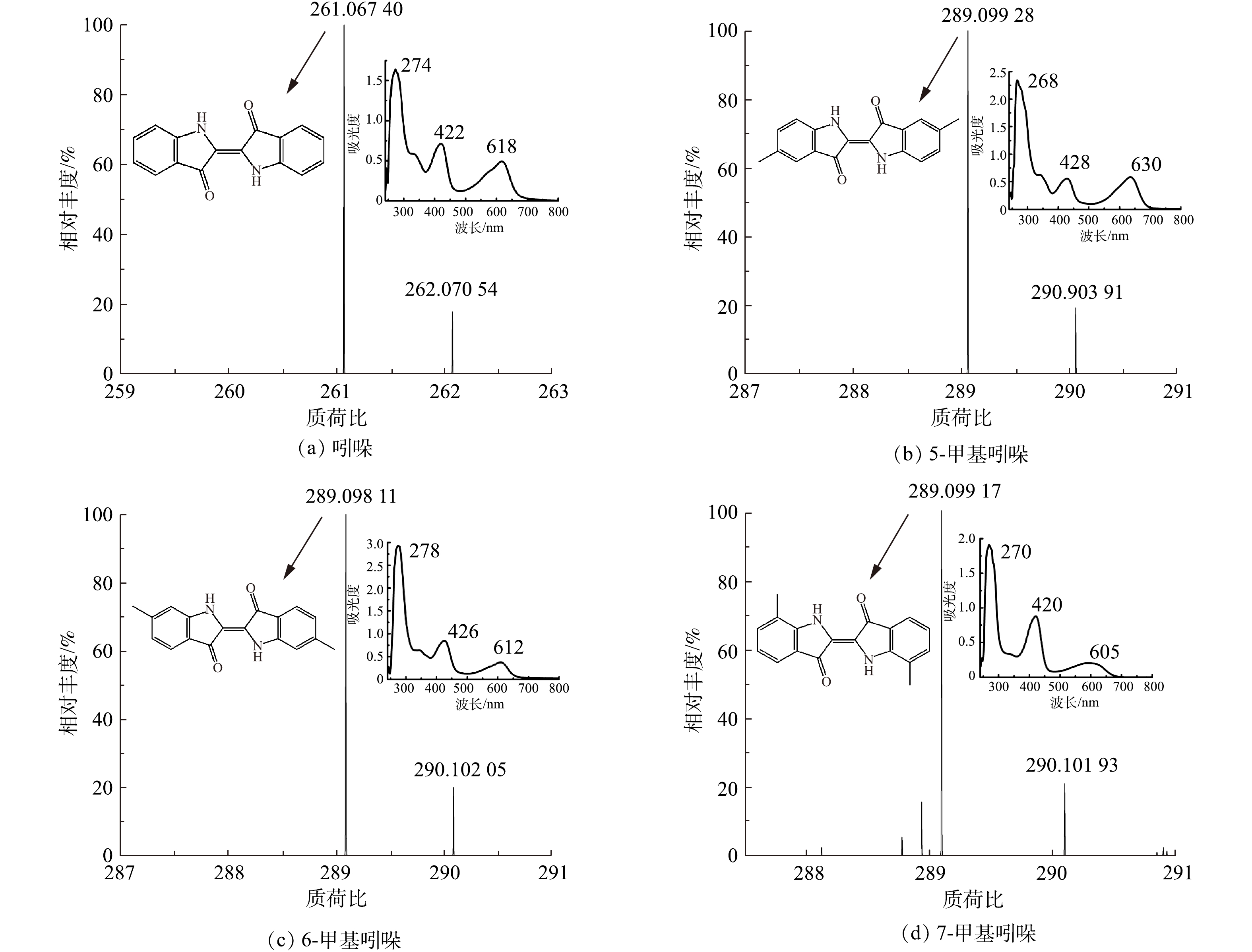
实验对吲哚、5-甲基吲哚、6-甲基吲哚和7-甲基吲哚转化生成的产物进行LC-MS分析,结果见图4。由图4(a)可知,吲哚的转化产物最大吸收波长为274、422和618 nm,且该蓝色产物的分子离子峰为261.067 40([M-H+]),与标准靛蓝样品的特征基本一致,由此推测菌株YC转化吲哚合成的产物为靛蓝。在图4(b)中,5-甲基吲哚的转化产物的最大吸收波长为268、428和630 nm,与靛蓝具有类似的特征且产物的分子离子峰为289.099 28([M-H+]),由此推测菌株YC转化5-甲基吲哚的产物可能为5,5′-二甲基靛蓝。类似地,6-甲基吲哚的转化产物最大吸收波长为278、426和612 nm,产物分子离子峰为289.098 11([M-H+]),其可能是6,6′-二甲基靛蓝(图4(c))。7-甲基吲哚的转化产物的最大吸收波长为270、420和605 nm,产物分子离子峰为289.099 17([M-H+]),产物可能为7,7′-二甲基靛蓝(图4(d))。已有研究[3, 17]对吲哚的转化产物开展了较多探讨,但对于吲哚衍生物的转化产物研究相对较少。本研究发现5-、6-、7-甲基吲哚都具有转化为靛蓝类色素的潜质。
-
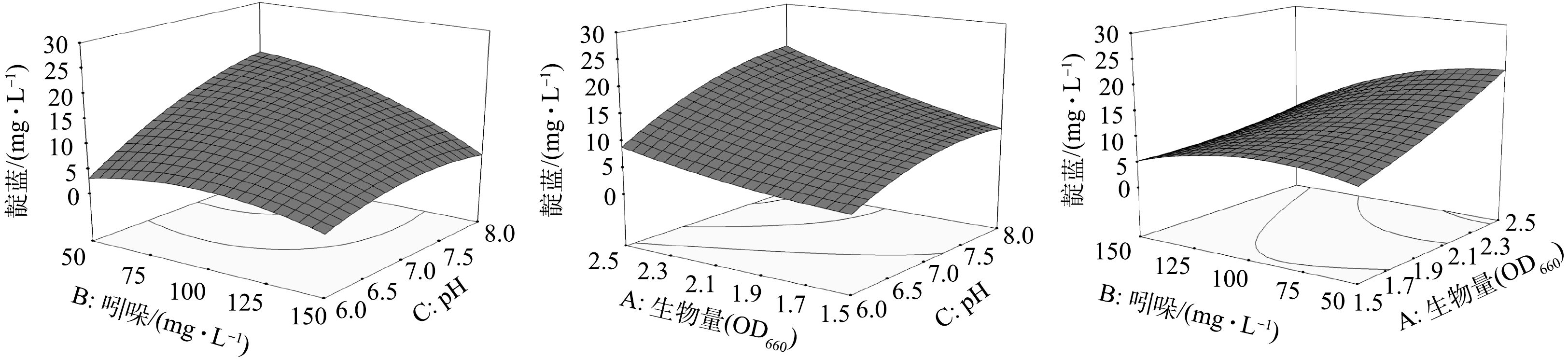
在吲哚的生物转化过程中,投加休眠菌体量与底物浓度对产物的合成应有较为直接的影响,而反应体系的pH则可能对细胞分泌的酶蛋白活性产生影响,进而影响细胞体转化底物的特性。因此,选取生物量(OD660,因素A)、吲哚浓度(因素B)和反应体系pH(因素C)为主要影响因素,以靛蓝产量为响应目标(Y),利用响应曲面法对菌株YC转化吲哚合成靛蓝的条件进行优化,结果如表1所示。利用Design Expert 8.0对实验数据进行拟合,得到了式(1)的预测值函数,并根据计算得到目标的预测响应值(表1)。
利用方差分析(ANOVA)对回归模型进行显著性检验,结果见表2。其中,模型的F值为14.22,P值小于0.05,说明模型具有显著性。此外,A、B、C、B2和C2的P值均小于0.05,说明生物量、吲哚浓度与反应体系pH对菌株YC转化吲哚合成靛蓝均具有显著影响。AB与BC的P值小于0.05,说明生物量与吲哚浓度之间、吲哚浓度与反应体系pH之间存在显著相关性。
利用Design Expert 8.0得到3个因素相互作用的3D响应图(图5)。由于吲哚是靛蓝合成的直接底物,在一定范围内,随着吲哚浓度的增加,靛蓝产率随之增加,但是吲哚对菌株及其体内加氧酶有一定的毒害作用,高浓度吲哚会降低吲哚的催化转化效率,从而抑制靛蓝的合成[18-19]。反应体系pH对吲哚转化酶的活性有重要影响,苯酚降解菌Pseudomonas sp. QM转化吲哚合成靛蓝的最适pH为7.2[14],萘降解菌Comamonas sp. MQ合成靛蓝的最适pH为6.9[20],而在本研究中,弱碱性条件(pH 8.00)更有利于菌株YC对吲哚进行生物转化合成靛蓝。
经模型预测,得到YC合成靛蓝的最优条件如下:生物量OD660为2.50,吲哚浓度为50.00 mg·L−1,反应体系pH为8.00。在此条件下,YC转化吲哚合成靛蓝的产量最大,为28.39 mg·L−1。
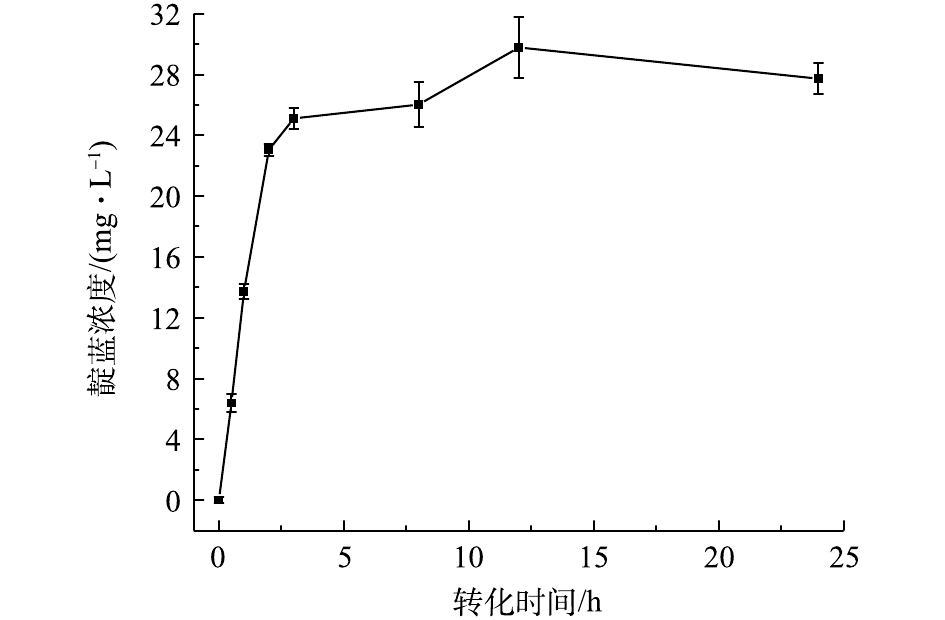
根据预测条件进行实验并绘制合成靛蓝的时间曲线(如图6所示)。在0~2 h,靛蓝产率较高,12 h后,靛蓝产量基本趋于稳定。靛蓝合成的最大值为29.78 mg·L−1,该值与模型预测值(28.39 mg·L−1)基本相当,这进一步证实了模型的可靠性。
在前期研究中,苯酚降解菌能够较好地转化吲哚合成靛蓝。QU等[13]分离得到的Pseudomonas monteilii QM在苯酚诱导下,合成靛蓝的产量为27.2 mg·L−1。WANG等[14]分离的苯酚降解菌Pseudomonas sp. P1和Acinetobacter sp. PI2在额外添加酵母粉的条件下,合成靛蓝的产量分别为11.82 mg·L−1和17.19 mg·L−1。而由胜男等[8]筛选的Comamonas sp. IDO2也可以在苯酚诱导下,转化吲哚合成靛蓝,产量为9.0 mg·L−1。与这些菌株相比,Pseudomonas sp. YC具有较好的吲哚转化合成靛蓝的能力。尽管如此,菌株YC转化吲哚的效率与产量仍相对偏低。在后续研究中,可以结合基因组学与比较蛋白质组学,探究菌株YC中与吲哚转化及靛蓝合成相关的功能基因,建立适宜的调控机制,以提高吲哚转化效率,从而为吲哚废水的资源化处理提供参考。
-
1)从土壤中分离得到具有降解苯酚能力的菌株YC,经16S rRNA基因序列分析比对,鉴定为Pseudomonas sp.。
2)菌株YC的休眠细胞能转化吲哚及甲基吲哚,合成不同的靛蓝类色素,转化吲哚、5-甲基吲哚、6-甲基吲哚和7-甲基吲哚的蓝色产物分别为靛蓝、5,5′-二甲基靛蓝、6,6′-二甲基靛蓝和7,7′-二甲基靛蓝。
3)利用响应曲面法考察了生物量、吲哚浓度和反应体系pH对菌株YC转化吲哚合成靛蓝的影响,得到靛蓝合成的最优化条件为:生物量OD660 2.50、吲哚浓度50.00 mg·L−1,反应体系pH 8.00。在此条件下,靛蓝产量为29.78 mg·L−1。
苯酚降解菌生物转化吲哚及甲基吲哚的特性
Characteristics of indole and methylindoles biotransformation by phenol degrading bacteria
-
摘要: 为探索吲哚类废水的资源化处置效果,考察了苯酚降解菌对吲哚及甲基吲哚的生物转化行为,从土壤中分离筛选苯酚降解菌,结合16S rRNA序列分析对菌株进行鉴定;利用菌株休眠细胞对吲哚及甲基吲哚进行生物转化,采用液相色谱-质谱联用分析其转化产物,并结合响应曲面法对吲哚转化合成靛蓝的条件进行优化。结果表明:筛选得到的1株苯酚降解菌YC为Pseudomonas菌属;菌株YC的休眠细胞可对吲哚、5-甲基吲哚、6-甲基吲哚、7-甲基吲哚进行生物转化,生成靛蓝及甲基取代靛蓝。菌株转化吲哚合成靛蓝的最适条件为:生物量OD660 2.50,吲哚50.00 mg·L−1,反应体系pH 8.00,在此条件下,靛蓝产量可达到29.78 mg·L−1。综合上述结果,苯酚降解菌Pseudomonas sp. YC能较好地转化吲哚及甲基吲哚合成靛蓝类色素,在吲哚类废水生物修复中具有一定的应用前景。Abstract: In this study, the biotransformation behavior of indole and methylindoles by phenol-degrading bacteria was explored, as well as the resource utilization of the indoles wastewater. The phenol-degrading bacterial strain was isolated from soil, then was identified by 16S rRNA sequence analysis. The indole and methylindoles were biotransformed by the resting cells of strain, and the products were analyzed by liquid chromatography-mass spectrometry (LC-MS). The response surface methodology (RSM) was used to optimize the conditions of indoles biotransformation and indigo blue preparation. The results showed that a phenol-degrading bacterial strain YC was isolated and identified as Pseudomonas sp. Indole, 5-methylindole, 6-methylindole and 7-methylindole could be transformed to indigo and methyl substituted indigo by the resting cells of strain YC. The optimal conditions for indole biotransformation to indigo were as follows: biomass OD660 of 2.50; indole of 50.00 mg·L−1 and pH 8.00. Under the optimal conditions, the indigo yield could reach 29.78 mg·L−1. This indicated that the phenol-degrading strain Pseudomonas sp. YC could transform indole and methylindoles to indigoid pigments, which presents a promising potential application in the bioremediation of indole wastewater.
-
Key words:
- phenol-degrading bacteria /
- indole /
- methylindoles /
- indigo /
- surface response methodology
-
吲哚常见于焦化废水中,是一种典型的氮杂环污染物。环境中过量的吲哚类物质会对人体造成伤害,故对吲哚废水的处理与资源化已成为丞待解决的问题[1-4]。研究表明,微生物能够对吲哚进行生物转化,生成靛蓝、靛玉红等具有高附加值的靛蓝类色素。相关研究有助于探索吲哚废水的资源化处置,近年来已逐渐受到研究者的关注[5-11]。
研究发现,苯酚降解菌及相关功能酶能够较好地转化吲哚合成靛蓝类色素。KIM等[12]从苯酚降解菌Pseudomonas sp. KL28中克隆表达的多组分苯酚羟化酶(mPHKL28)能够转化吲哚,合成靛蓝和7-羟基吲哚,也能转化多种吲哚衍生物,合成具有不同颜色的靛蓝类色素。QU等[13]分离得到的苯酚降解菌Pseudomonas monteilii QM可转化吲哚,合成靛蓝、靛玉红等物质,WANG等[14]筛选的Pseudomonas sp. PI1和Acinetobacter sp. PI2也可在苯酚诱导下转化吲哚,合成多种靛蓝类色素。目前,苯酚降解菌对吲哚的生物转化研究报道较多,但是对于其他吲哚类物质(如甲基吲哚)的生物转化研究相对较少。
本研究在土壤中分离得到1株苯酚降解菌YC,通过形态观察及16S rRNA序列分析对菌株进行鉴定,考察菌株对吲哚及甲基吲哚的生物转化特性,并利用响应曲面法对菌株转化吲哚合成靛蓝的条件进行优化,为焦化废水中吲哚的生物处理与资源化提供高效的微生物资源。
1. 材料与方法
1.1 培养基
采用无机盐培养基,其组成包括Na2HPO4·12H2O 3.28 mg·L−1、KH2PO4 2 mg·L−1、(NH4)2SO4 2 mg·L−1、FeCl3 0.25 mg·L−1,pH=7.0。
1.2 试剂与仪器
实验所用吲哚、甲基吲哚、靛蓝、二甲基亚砜及无机盐培养基成分均为分析纯,甲醇为色谱纯,土壤样品取自大连理工大学盘锦校区校园。
使用的主要仪器包括:场发射扫描电子显微镜(Nova NanoSEM 450,美国FEI公司);紫外-可见分光光度计(UH5300,日本HITACHI公司);高效液相色谱仪(1290 Infinity II,德国Agilent公司);四级杆轨道离子阱高分辨质谱仪(Q Exactive,美国Thermo Fisher科技有限公司)。
1.3 菌株的分离与鉴定
取10 g土壤样品加入100 mL含苯酚(100 mg·L−1)的无机盐培养基,好氧培养7 d后,按体积分数10%接种量转接到新的无机盐培养基(含苯酚100 mg·L−1)继续培养。经3~4次培养后,取驯化后的样品反复进行平板涂布,最后得到具有苯酚降解能力的单菌落,命名为YC。利用扫描电子显微镜观察菌株的细胞形态。
以菌株YC菌落为模板,选择16S rRNA通用引物对其16S rRNA基因序列进行PCR扩增,送至生工生物工程(上海)股份有限公司检测,测序结果经Blast在Genbank中比对分析。利用MEGA 6.0软件构建系统发育树,确定菌株的分类。
1.4 菌株的培养与收集
采用含3 g·L−1酵母浸粉与100 mg·L−1苯酚的无机盐培养基进行富集培养,好氧培养24 h后,离心收集菌体(10 000 r·min−1,10 min),使用无机盐培养基洗涤2次后备用。
1.5 吲哚及甲基吲哚的转化实验
将收集好的菌体用无机盐培养基重悬,充分摇匀至无固形物,采用紫外-可见分光光度计测定菌悬液OD660,并利用无机盐培养基调节OD660为2.0。移取20 mL菌悬液分别加入50 mg·L−1吲哚或甲基吲哚,好氧培养24 h,取反应液进行后续分析。
1.6 靛蓝合成的条件优化
采用响应曲面法对菌株YC转化吲哚合成靛蓝进行条件优化。响应曲面法可通过建模分析,在最低限度的实验次数下,利用最小二分法对多个受变量影响的因素进行分析,并探究因素间的影响关系[15]。本研究以靛蓝产量为响应目标,探究生物量(因素A)、吲哚浓度(因素B)与反应体系pH(因素C)对生物转化过程的影响。采用Design Expert 8.0软件设计实验并进行分析,生物量为1.5~2.5(OD660),吲哚浓度为50~150 mg·L−1,pH为6~8,设计条件如表1所示,其中标号为15、16、17、18、19和20的实验组为各因素中心点值的实验组合。
表 1 吲哚的生物转化合成靛蓝的条件优化与结果Table 1. Condition optimization of indole biotransformation and indigo preparation and the corresponding experimental results编号 生物量/(OD660) 吲哚/(mg·L−1) pH 靛蓝/(mg·L−1) 实验值 预测值 1 1.50 50.00 6.00 1.37 0.00 2 2.50 50.00 6.00 6.67 11.19 3 1.50 150.00 6.00 0.24 2.24 4 2.50 150.00 6.00 1.72 5.61 5 1.50 50.00 8.00 11.96 11.23 6 2.50 50.00 8.00 27.50 28.67 7 1.50 150.00 8.00 3.54 2.19 8 2.50 150.00 8.00 4.94 10.64 9 1.16 100.00 7.00 7.63 10.71 10 2.84 100.00 7.00 35.76 28.20 11 2.00 15.91 7.00 9.15 9.24 12 2.00 184.09 7.00 1.50 0.00 13 2.00 100.00 5.32 0.43 0.00 14 2.00 100.00 8.68 13.27 11.94 15 2.00 100.00 7.00 13.70 12.74 16 2.00 100.00 7.00 10.52 12.74 17 2.00 100.00 7.00 15.47 12.74 18 2.00 100.00 7.00 12.73 12.74 19 2.00 100.00 7.00 11.11 12.74 20 2.00 100.00 7.00 12.13 12.74 1.7 产物的分离及鉴定
1) 液相色谱(HPLC)分析。取反应液离心,沉淀加入适量二甲基亚砜重悬离心,取上清液过0.45 μm有机膜后进行HPLC分析。HPLC条件如下:Hypersil ODS2 C18色谱柱(5 μm,4.6 × 250 mm),流速1.0 mL·min−1,进样量5 μL,流动相为水/甲醇;洗脱时间共计20 min,0~15 min进行梯度洗脱,甲醇含量由60%升至70%;15~20 min进行等度洗脱,甲醇含量60%。
2) 液相-质谱(LC-MS)分析。产物分离过程同上,LC条件:Ultimate XB-C18色谱柱(3 μm, 2.1 × 150 mm),流速0.4 mL·min−1,进样量10 μL,流动相为水/甲醇;洗脱时间共计5 min,0~2 min进行梯度洗脱,甲醇含量由50%升至95%;2~4 min进行等度洗脱,甲醇含量95%;4~5 min进行梯度洗脱,甲醇含量50%。MS条件:采用ESI离子源,雾化室温度320 ℃,以N2为保护气和干燥气,进行负模式扫描。
2. 结果与讨论
2.1 苯酚降解菌YC的分离与鉴定
通过反复平板涂布,筛选得到了1株能以苯酚为唯一碳源和能源生长的菌株,命名为YC。菌株YC在固体培养基上的菌落呈白色,为表面光滑且边缘规则的圆形,扫描电镜下为杆状(图1)。通过16S rRNA序列对比发现,菌株YC的16S rRNA(GenBank登录号MH185838)与多株Pseudomonas属菌株的序列相似度为98%以上,因此,菌株YC为Pseudomonas sp.(假单胞菌属)。采用比邻法构建菌株YC的系统发育树(如图2所示),结果显示,菌株YC位于Pseudomonas属簇内,并且与Pseudomonas plecoglossicida菌种及Pseudomonas monteilii菌种的亲缘性更近。
2.2 苯酚降解菌YC对吲哚及甲基吲哚的生物转化
预实验结果表明,菌株YC在含有苯酚(100 mg·L−1)与吲哚(50 mg·L−1)的无机盐培养基中生长时,能够生成蓝色的气泡,表明可能有靛蓝类色素合成,但菌株生长量较低,靛蓝产量也相对较低。因此,考虑利用添加了酵母浸粉和苯酚的无机盐培养基对菌株进行富集培养,其中酵母浸粉能够为菌株生长提供充足的营养物质,而苯酚能够诱导菌株表达芳烃加氧酶(如苯酚羟化酶)。将培养好的菌株离心收集,并通过无机盐培养基重悬制备休眠细胞,选取吲哚、2-甲基吲哚、3-甲基吲哚、5-甲基吲哚、6-甲基吲哚、7-甲基吲哚,考察菌株YC休眠细胞生物转化吲哚及甲基吲哚合成靛蓝类色素的可行性。由图3可知,菌株YC能够较好地转化吲哚(6)、5-甲基吲哚(3)、6-甲基吲哚(4)、7-甲基吲哚(5)生产蓝色或紫色的产物,但2-甲基吲哚(1)的转化产物为黄色,而3-甲基吲哚(4)的反应后无明显颜色。
吲哚可经氧化生成2-羟吲哚和3-羟吲哚,靛蓝则常由3-羟吲哚二聚形成[3]。相较其他甲基吲哚,2-甲基吲哚由于其2号碳原子位置上的甲基极有可能阻碍苯酚羟化酶在3号碳位置上的加氧羟化,从而阻碍靛蓝类色素的合成,故其并未显著合成靛蓝类色素[12, 16]。而3-甲基吲哚的3号碳位置上的甲基则会完全阻碍了3号碳位置上的羟化,由此推断,其无法通过在3号碳位置上进行羟化得到靛蓝类色素。实验结果表明苯酚降解菌YC具有较为广谱的靛蓝类色素合成能力。
2.3 吲哚及甲基吲哚生物转化产物的鉴定
对吲哚、5-甲基吲哚、6-甲基吲哚和7-甲基吲哚转化生成的产物进行LC-MS分析。由图4(a)可知,吲哚的转化产物最大吸收波长为274、422和618 nm,且该蓝色产物的分子离子峰为261.067 40([M-H+]),与标准靛蓝样品的特征基本一致,由此推测菌株YC转化吲哚合成的产物为靛蓝。在图4(b)中,5-甲基吲哚的转化产物的最大吸收波长为268、428和630 nm,与靛蓝具有类似的特征且产物的分子离子峰为289.099 28([M-H+]),由此推测菌株YC转化5-甲基吲哚的产物可能为5,5′-二甲基靛蓝。类似地,6-甲基吲哚的转化产物最大吸收波长为278、426和612 nm,产物分子离子峰为289.098 11([M-H+]),其可能是6,6′-二甲基靛蓝(图4(c));7-甲基吲哚的转化产物的最大吸收波长为270、420和605 nm,产物分子离子峰为289.099 17([M-H+]),产物可能为7,7′-二甲基靛蓝(图4(d))。已有文献对吲哚的转化产物开展了较多研究,但对于吲哚衍生物的转化产物研究相对较少[3, 17]。本研究发现5-、6-、7-甲基吲哚都具有转化为靛蓝类色素的潜质。
2.4 吲哚及甲基吲哚生物转化产物的鉴定
在吲哚的生物转化过程中,投加休眠菌体量与底物浓度对产物的合成应有较为直接的影响,而反应体系的pH则可能对细胞分泌的酶蛋白活性产生影响,进而影响细胞体转化底物的特性。因此,选取生物量(OD660,因素A)、吲哚浓度(mg·L−1,因素B)和反应体系pH(因素C)为主要影响因素,以靛蓝产量为响应目标(Y),利用响应曲面法对菌株YC转化吲哚合成靛蓝的条件进行优化,结果如表1所示。利用Design Expert 8.0对实验数据进行回归拟合,得到了式(1)的预测值函数,并根据计算得到目标的预测响应值(表1)。
Y=−173.7−36.38A+0.8152B+45.73C−8.989×10−2AB+2.540AC−6.223×10−2BC+9.499A2−1.364×10−3B2−2.874C2 (1) 利用方差分析(ANOVA)对回归模型进行显著性检验,结果见表2。其中,模型的F值为14.22,P值小于0.05,说明模型具有显著性。此外,A、B、C、B2和C2的P值均小于0.05,说明生物量、吲哚浓度与反应体系pH对菌株YC转化吲哚合成靛蓝均具有显著影响。AB与BC的P值小于0.05,说明生物量与吲哚浓度之间、吲哚浓度与反应体系pH之间存在着显著的相互影响。
表 2 模型中不同因素的ANOVA结果Table 2. ANOVA results for different factors in model因素 偏差平法和 均方和 F值 P值1) 模型 1 093.64 121.52 14.22 0.000 1 A 220.69 220.69 25.83 0.000 5 B 203.86 203.86 23.86 0.000 6 C 284.82 284.82 33.34 0.000 2 AB 54.17 54.17 6.34 0.030 5 AC 21.12 21.12 2.47 0.140 7 BC 96.13 96.13 11.25 0.007 3 A2 11.06 11.06 1.29 0.281 8 B2 122.48 122.48 14.34 0.003 6 C2 81.48 81.48 9.54 0.011 5 注:1)P < 0.05则说明模型具有显著性。 利用Design Expert 8.0得到3个因素相互作用的3D响应图(图5)。由于吲哚是靛蓝合成的直接底物,故在一定范围内,随着吲哚浓度的增加,靛蓝产率随之增加,但是吲哚对菌株及其体内加氧酶有一定的毒害作用,高浓度吲哚会降低吲哚的催化转化效率,从而抑制靛蓝的合成[18-19]。反应体系pH对吲哚转化酶的活性有重要影响,苯酚降解菌Pseudomonas sp. QM转化吲哚合成靛蓝的最适pH为7.2[14],萘降解菌Comamonas sp. MQ合成靛蓝的最适pH为6.9[20],而在本研究中,弱碱性条件(pH 8.00)更有利于菌株YC对吲哚进行生物转化合成靛蓝。
经模型预测,得到了YC合成靛蓝的最优条件如下:生物量OD660 2.50、吲哚浓度50.00 mg·L−1,反应体系pH 8.00。在此条件下,YC转化吲哚合成靛蓝的产量最大,为28.39 mg·L−1。
根据预测条件进行实验并绘制合成靛蓝的时间曲线(如图6所示),在0~2 h内,靛蓝产率较高,12 h后,靛蓝产量基本趋于稳定。靛蓝合成的最大值为29.78 mg·L−1,该值与模型预测值(28.39 mg·L−1)基本相当,进一步证实模型的可靠性。
在前期研究中,苯酚降解菌能够较好地转化吲哚合成靛蓝。QU等[13]分离得到的Pseudomonas monteilii QM在苯酚诱导下,合成靛蓝的产量为27.2 mg·L−1;WANG等[14]分离的苯酚降解菌Pseudomonas sp. P1和Acinetobacter sp. PI2在额外添加酵母粉的条件下,合成靛蓝的产量分别为11.82 mg·L−1和17.19 mg·L−1;而由胜男等[8]筛选的Comamonas sp. IDO2也可以在苯酚诱导下,转化吲哚合成靛蓝,产量为9.0 mg·L−1。与这些菌株相比,Pseudomonas sp. YC具有较好的吲哚转化合成靛蓝的能力。尽管如此,菌株YC转化吲哚的效率与产量仍相对偏低。在后续研究中,可以结合基因组学与比较蛋白质组学,探究菌株YC中与吲哚转化及靛蓝合成相关的功能基因,建立适宜的调控机制,以提高吲哚转化效率,从而为吲哚废水的资源化处理提供参考。
3. 结论
1)从土壤中分离得到具有降解苯酚能力的菌株YC,经16S rRNA基因序列分析比对,鉴定为Pseudomonas sp.。
2)菌株YC的休眠细胞能转化吲哚及甲基吲哚合成不同的靛蓝类色素,转化吲哚、5-甲基吲哚、6-甲基吲哚和7-甲基吲哚的蓝色产物分别为靛蓝、5,5′-二甲基靛蓝、6,6′-二甲基靛蓝和7,7′-二甲基靛蓝。
3)利用响应曲面法考察了生物量、吲哚浓度和反应体系pH对菌株YC转化吲哚合成靛蓝的影响,得到靛蓝合成的最优化条件为:生物量OD660 2.50、吲哚浓度50.00 mg·L−1,反应体系pH 8.00;在此条件下,靛蓝产量为29.78 mg·L−1。
-
表 1 吲哚的生物转化合成靛蓝的条件优化与结果
Table 1. Condition optimization of indole biotransformation and indigo preparation and the corresponding experimental results
编号 生物量(OD660) 吲哚/(mg·L−1) pH 靛蓝/(mg·L−1) 实验值 预测值 1 1.50 50.00 6.00 1.37 0 2 2.50 50.00 6.00 6.67 11.19 3 1.50 150.00 6.00 0.24 2.24 4 2.50 150.00 6.00 1.72 5.61 5 1.50 50.00 8.00 11.96 11.23 6 2.50 50.00 8.00 27.50 28.67 7 1.50 150.00 8.00 3.54 2.19 8 2.50 150.00 8.00 4.94 10.64 9 1.16 100.00 7.00 7.63 10.71 10 2.84 100.00 7.00 35.76 28.20 11 2.00 15.91 7.00 9.15 9.24 12 2.00 184.09 7.00 1.50 0 13 2.00 100.00 5.32 0.43 0 14 2.00 100.00 8.68 13.27 11.94 15 2.00 100.00 7.00 13.70 12.74 16 2.00 100.00 7.00 10.52 12.74 17 2.00 100.00 7.00 15.47 12.74 18 2.00 100.00 7.00 12.73 12.74 19 2.00 100.00 7.00 11.11 12.74 20 2.00 100.00 7.00 12.13 12.74 表 2 模型中不同因素的ANOVA结果
Table 2. ANOVA results for different factors in model
因素 偏差平法和 均方和 F值 P值1) 模型 1 093.64 121.52 14.22 0.000 1 A 220.69 220.69 25.83 0.000 5 B 203.86 203.86 23.86 0.000 6 C 284.82 284.82 33.34 0.000 2 AB 54.17 54.17 6.34 0.030 5 AC 21.12 21.12 2.47 0.140 7 BC 96.13 96.13 11.25 0.007 3 A2 11.06 11.06 1.29 0.281 8 B2 122.48 122.48 14.34 0.003 6 C2 81.48 81.48 9.54 0.011 5 注:1)P < 0.05则说明模型具有显著性。 -
[1] GUY R H, JEFFREY T K. Practical methodologies for the synthesis of indoles[J]. Chemical Reviews, 2006, 106(7): 2875-2911. doi: 10.1021/cr0505270 [2] 徐兆瑜. 医药化工中的一些重要吲哚化合物[J]. 精细化工原料及中间体, 2009(1): 27-30. [3] MA Q, ZHANG X, QU Y. Biodegradation and biotransformation of indole: Advances and perspectives[J]. Frontiers in Microbiology, 2018, 9: 2625. doi: 10.3389/fmicb.2018.02625 [4] QU Y, SHEN E, MA Q, et al. Biodegradation of indole by a newly isolated Cupriavidus sp. SHE[J]. Journal of Environmental Sciences, 2015, 34(8): 126-132. [5] 韩晓红, 王伟, 肖兴国. 靛蓝及其同类色素的微生物生产与转化[J]. 生物工程学报, 2008, 24(6): 921-926. doi: 10.3321/j.issn:1000-3061.2008.06.003 [6] 马桥, 曲媛媛, 张旭旺, 等. 靛蓝的微生物合成研究新进展[J]. 应用与环境生物学报, 2012, 18(2): 344-350. [7] BOYD C, LARKIN M J, REID K A, et al. Metabolism of naphthalene, 1-naphthol, indene, and indole by Rhodococcus sp. strain NCIMB 12038[J]. Applied & Environmental Microbiology, 1997, 63(1): 151-155. [8] MURDOCK D, ENSLEY B D, SERDAR C, et al. Construction of metabolic operons catalyzing the de novo biosynthesis of indigo in Escherichia coli[J]. Biotechnology, 1993, 11(3): 381-386. doi: 10.1038/nbt0393-381 [9] GUI H H, SHIN H J, SI W K. Optimization of bio-indigo production by recombinant E. coli, harboring fmo gene[J]. Enzyme & Microbial Technology, 2008, 42(7): 617-623. [10] HAN G H, BANG S E, BABU B K, et al. Bio-indigo production in two different fermentation systems using recombinant[J]. Process Biochemistry, 2011, 46(3): 788-791. doi: 10.1016/j.procbio.2010.10.015 [11] 由胜男, 沈文丽, 裴晓芳, 等. Comamonas sp. IDO2合成靛蓝的特性研究[J]. 环境科学学报, 2017, 37(6): 2069-2075. [12] KIM J Y, KIM J K, LEE S O, et al. Multicomponent phenol hydroxylase-catalysed formation of hydroxyindoles and dyestuffs from indole and its derivatives[J]. Letters in Applied Microbiology, 2005, 41: 163-168. doi: 10.1111/lam.2005.41.issue-2 [13] QU Y, MA Q, ZHANG X, et al. Optimization of indigo production by a newly isolated Pseudomonas sp. QM[J]. Journal of Basic Microbiology, 2012, 52(6): 687-694. doi: 10.1002/jobm.v52.6 [14] WANG J, ZHANG X, FAN J, et al. Indigoids biosynthesis from indole by two phenol-degrading strains, Pseudomonas sp. PI2[J]. Applied Biochemistry and Biotechnology, 2015, 176(5): 1263-1276. doi: 10.1007/s12010-015-1644-9 [15] 王永菲, 王成国. 响应面法的理论与应用[J]. 中央民族大学学报(自然科学版), 2005, 14(3): 236-240. doi: 10.3969/j.issn.1005-8036.2005.03.008 [16] KIM J Y, LEE K, KIM Y, et al. Production of dyestuffs from indole derivatives by naphthalene dioxygenase and toluene dioxygenase[J]. Letters in Applied Microbiology, 2003, 36(6): 343-348. doi: 10.1046/j.1472-765X.2003.01279.x [17] O′CONNOR K E, HARTMANS S. Indigo formation by aromatic hydrocarbon-degrading bacteria[J]. Biotechnology Letters, 1998, 20(3): 219-223. doi: 10.1023/A:1005361415496 [18] LU Y, MEI L. Optimization of fermentation conditions for P450 BM-3 monooxygenase production by hybrid design methodology[J]. Journal of Zhejiang University Science B, 2007, 8(1): 27-32. doi: 10.1631/jzus.2007.B0027 [19] DOUKYU N, NAKANO T, OKUYAMA Y, et al. Isolation of an Acinetobacter sp. ST-550 which produces a high level of indigo in a water-organic solvent two-phase system containing high levels of indole[J]. Applied Microbiology and Biotechnology, 2002, 58(4): 543-546. doi: 10.1007/s00253-001-0919-y [20] 李诗阳, 袁琪, 黄益养, 等. 萘降解菌MQ合成靛蓝的研究[J]. 环境科学学报, 2012, 32(8): 1807-1813. 期刊类型引用(1)
1. 陈卓,郭鑫宇,吴炜泽,戴春晓,荆佳维,曲媛媛. 芳烃诱导Acinetobacter sp. CZ合成靛蓝及染布应用研究. 环境科学学报. 2023(10): 37-46 .  百度学术
百度学术
其他类型引用(1)
-






 DownLoad:
DownLoad: