-
阿特拉津(2-Chloro-4-ethylamino-6-isopropylamino-s-triazine,ATZ)是一种通过阻断叶绿体中质体醌结合蛋白和抑制光合作用来防治阔叶和禾草类杂草的选择性除草剂,土壤对其吸附性较低,因此极易向地表水、深层土壤和地下中迁移[1]. 低浓度的阿特拉津显著降低作物的株高、根长、根干重,高浓度的阿特拉津甚至会导致植物死亡[2];阿特拉津影响雄性斑马鱼的神经发育和神经功能[3],已成为一种污染源并对动植物和人类健康造成不可忽视的风险[4-5]. 由于阿特拉津严重的危害及影响,2004年在欧盟被禁止使用,但由于它价格低廉而在世界其他地区被广泛使用,仍是国际上销售的主要除草剂之一.
目前,环境中阿特拉津的去除方法主要有吸附法[6]、光催化法[7]、高级氧化法[8]、植物修复法[9-10]、生物法[11]等,其中基于微生物降解的生物修复法不仅效率高,而且几乎不损害生态环境,并且可将ATZ转化为无毒或低毒的物质.很多研究者已筛选出能将阿特拉津作为唯一氮源或碳源的微生物,如细菌Pseudomonas sp. strain AKN5[12]、Klebsiella sp. FH-1[13]、Citricoccus sp. strain TT3[14]、Arthrobacter sp. 30、Pseudomonas sp. AD39[15]和真菌Pleuroyus ostreatus INCQS 40310[16]等. 这些菌株对阿特拉津具有耐受性并能降解高浓度的阿特拉津,Enterobacter sp.LY-2[17]对浓度为100 mg·kg−1的污染土壤具有修复效果,14 d后阿特拉津浓度降低为9.9 mg·kg−1;产脲节杆菌(Arthrobacter ureafaciens)CS3,培养2 d 可将50 mg·L−1的阿特拉津完全降解[18]. 虽然目前报道的去除阿特拉津的微生物种类较多,但仍然存在停滞期长,降解耗时较长,耐受性差等不足。而且大部分菌株来源于中国北部寒冷地区,东北多为黑土,呈中性或偏碱性、有机质及氮磷含量丰富,而长三角地区雨量充沛、气温较高,土壤通透性差、呈弱酸性、有机质及氮磷含量较低,这些菌株可能不适应长三角的土壤环境,因此有必要寻找适合长三角地区的高效阿特拉津降解微生物资源.
本研究从江苏省5个不同地区的农田土壤中筛选分离能适应长三角地区阿特拉津污染土壤的菌株,研究ATZ初始浓度、温度和pH对菌株繁殖和降解效果的影响,判断菌株的适用环境范围,并采用HPLC-MS测定ATZ的降解产物,推断可能存在的降解转化途径.
-
阿特拉津(>97%)(C18H14ClN5,分子量214.69,密度为1.2 g·cm−1、沸点为200 ℃、熔点为174 ℃,难溶于水)、甲醇、乙醇、葡萄糖、C6H9Na3O9、C9H11NO、C6H7NO3S、C10H9N等. 无机盐培养基(MSM):Mg2SO4·7H2O 0.1 g·L−1、K2HPO4 1 g·L−1、KH2PO4 1 g·L−1、FeSO4·7H2O 0.025 g·L−1、CaCl2 0.025 g·L−1,适量阿特拉津;富集培养基(LB) :牛肉浸膏5 g·L−1、胰蛋白胨10 g·L−1、NaCl 5 g·L−1,加15 g·L−1琼脂即为固体培养基.
-
取江苏省5个长期喷洒阿特拉津的农田表层土壤(0—10 cm)进行降解菌株的驯化. 将土壤样品(5 g)分别加入150 mL MSM培养基中(ATZ浓度为50 mg·L−1),培养条件设置为 30 ℃、150 r·min−1. 驯化7 d后将上清液转至新的MSM培养基中,重复3次适应过程. 最后,将梯度稀释的培养液涂布于LB固体培养基,待菌落长成后,将其进行多次划线分离和纯化,并于无机盐培养基验证降解效果. 最终,筛选出一株能够降解阿特拉津的菌株D2,并采用斜面培养基和甘油保存.
-
将菌株接种于固体培养基,2—3 d后观察长出菌落的形态(形状、色泽、透明程度等);采用扫描电镜(Quanta FEG 250)观察菌株 D2放大50000倍后的表面形态.
-
革兰氏染色试验、水解产酸、柠檬酸盐利用、V-P、过氧化氢酶等指标,具体测定方法依据《污染控制微生物实验》[19],并参照《常见细菌系统鉴定手册》[20]与类似细菌进行比较.
-
将菌株在固体培养基中划线分离,待菌落长成后进行16S rDNA基因鉴定,由上海天霖生物科技有限公司完成. 将D2的 16Sr DNA 基因序列在BLAST系统中与基因库进行比对分析,并使用MEGA 7.0软件绘制系统发育树.
-
采用紫外可见分光光度计(INESA-N4)测定菌株生长不同时间段的OD600以推测菌株的生长情况,以原始溶液为空白对照绘制D2的生长曲线,并用 SGompertz模型[21](式(1))和Slogistic模型[22](式(3))拟合D2的生长曲线,并得到菌株最大比生长速率(μ1、μ2):
式中,t为时间(h),Nt1为时间t时菌株的数量,N0为初始菌株数量,Xc为达到最大升值速率的时间(h),μ1为菌株的最大比生长速率(h−1),a1为菌株最大生长量.
式中,t为时间(h),Nt2为时间t时菌株的数量,X0为达到最大升值速率的时间(h),μ2为菌株的最大比生长速率(h−1),a2为菌株最大生长量.
-
为了探究环境因素对菌株降解阿特拉津的影响,将菌悬液按照体积比为2%的量接种到含阿特拉津的MSM培养基中,研究不同初始浓度(10、20、50、100、150、200 mg·L−1)、不同pH(5.0、7.0、9.0)、不同温度(10、20、30、40 ℃)下菌株对阿特拉津的降解情况,以转速 150 r·min−1 避光培养,溶液经0.22 μm滤膜后测定阿特拉津的浓度.
-
阿特拉津的浓度采用岛津高效液相色谱仪(LC-20AT,Japan)测定. 检测条件:流动相为V(甲醇):V(水)=60:40,流速为1.0 mL·min−1,波长为220 nm. 降解产物定性分析采用带有高效液相色谱仪UltiMate 3000(Thermo Fisher Scientific,USA)和高分辨质谱仪5600 QTOF(AB SCIEX,Framingham,USA)的超高压液相质谱仪HPLC-MS,色谱柱(ACQUITY UPLC HSS T3 1.8 μm ,2.1 mm×100 mm),以水(含有2 mmol·L−1乙酸铵)和乙腈进行梯度洗脱. 条件:轰击能量,30 eV;雾化气压(GS1):60 Psi,辅助气压,60 Psi;气帘气压,35 Psi;温度,650 ℃;喷雾电压,5000 V(正离子模式)或−4000 V(负离子模式).
-
采用以阿特拉津(50 mg·L−1)为碳源和氮源的无机盐培养基对5个种植玉米的农田土壤经过驯化、分离,筛选出8株生长良好且菌落形态不同的菌株,最终选择一株降解效能最好的菌株,将其命名为D2.
菌株D2的菌落形态观察如图1(a),菌落较小,边缘整齐,浅黄色;革兰氏染色结果表明,D2为革兰氏阳性菌 (图1b ). 扫描电镜(×5000)下观察的菌株形态如图1(c)所示,D2呈杆状. 生理生化试验结果显示(表1),硝酸盐还原及过氧化氢酶试验均呈阳性,其余结果均为阴性.
将PCR扩增后得到的基因片段进行测序,测序结果在 NCBI 上经 BLAST 分析目标序列与同源序列,并绘制系统发育树(图2),对比发现ATZ降解菌株D2与土壤芽孢杆菌(Solibacillus)的核苷酸序列相似率高达99.58%,因此菌株D2经鉴定为土壤芽孢杆菌.
-
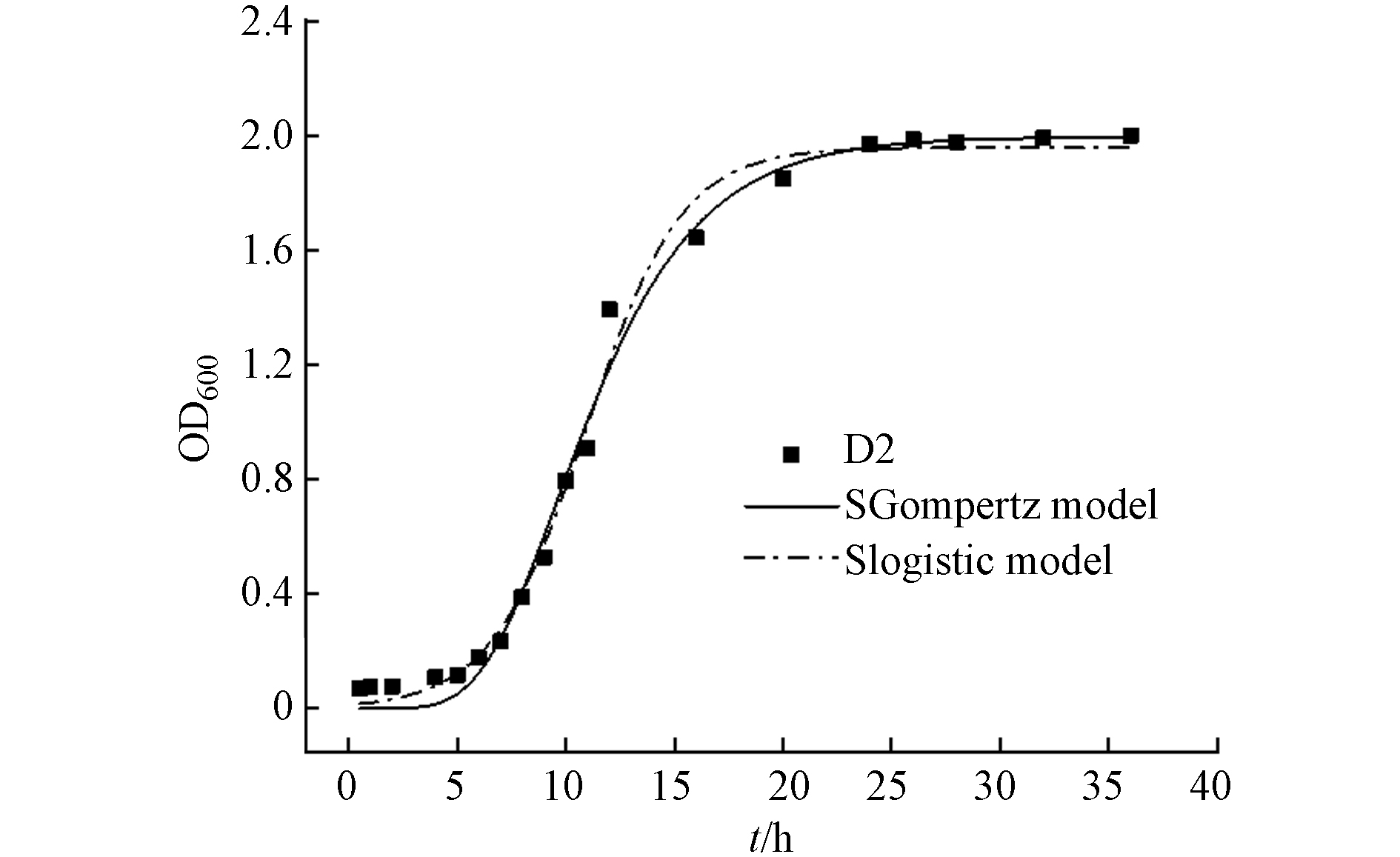
菌株D2的生长曲线见图3. 在LB培养基中,D2经过5 h的适应期后进入菌株代谢旺盛的对数期;12 h后OD600达到1.0,此时的菌株增长速率最快;24 h后达到生长繁殖的峰值,此后OD600不再增长,菌株数量相对趋于稳定.
采用SGompertz模型[23]和Slogistic模型拟合菌株的生长曲线(表2,图3). 结果显示,两种模型均可较好地拟合D2的生长曲线,拟合度(R2)分别为0.991和0.992,在第10 h左右D2到达最大比生长速率,随后比生长速率逐渐降低.
-
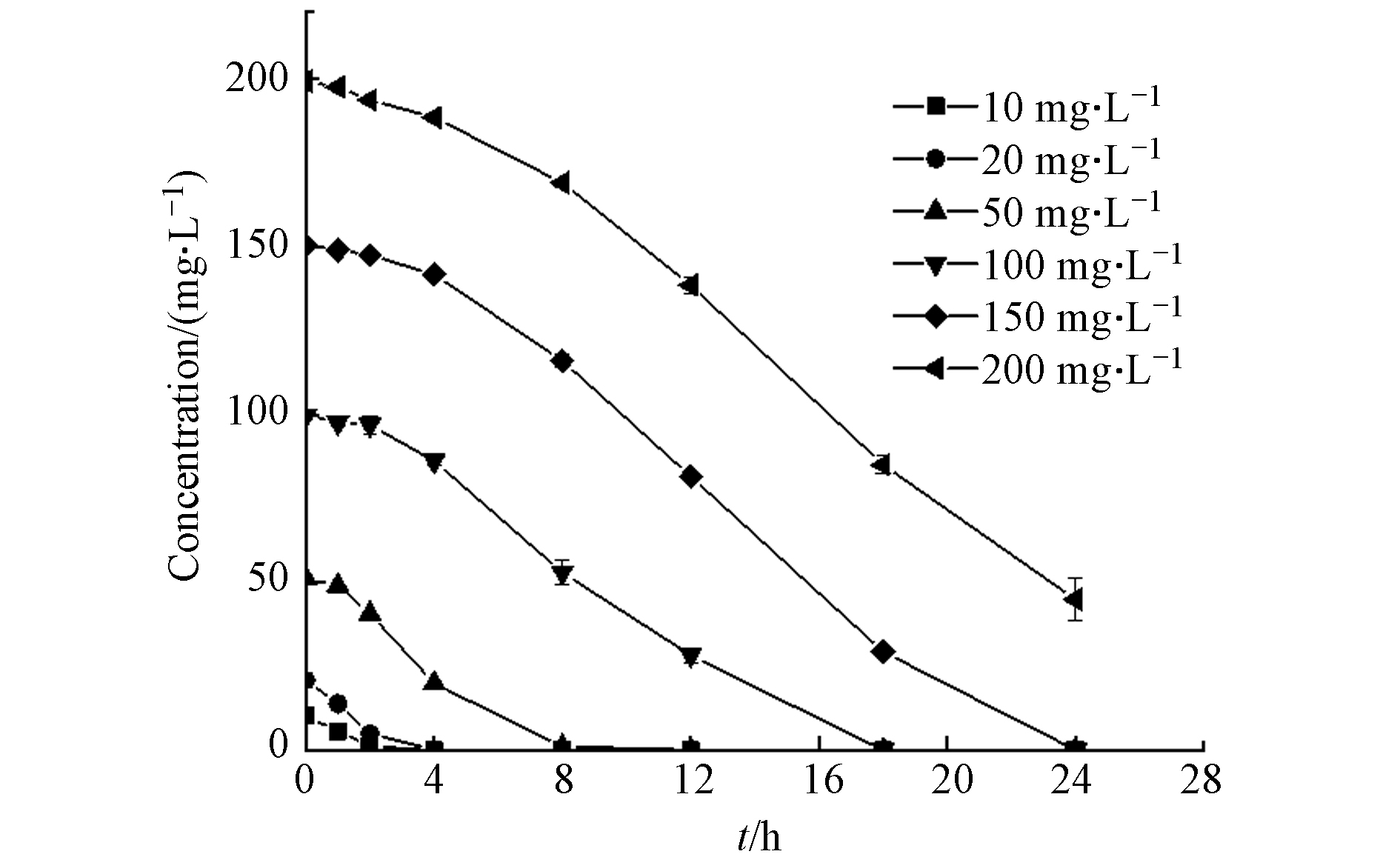
污染物浓度对菌株的影响是评价菌株生长及降解能力的标准之一[24],D2在不同初始浓度下对ATZ的降解效果如图4所示. 当浓度为200 mg·L−1时,D2对ATZ仍具有较高的去除能力,24 h的降解率为77.43%;初始浓度越低,D2的去除效率越高,将初始浓度为150、100、50、20、10 mg·L−1的ATZ完全降解分别需要24、18、8、4、3 h.长三角洲地区农田土壤中ATZ浓度的平均值为5.7 ng·L−1,检出率高达57.7%[25],菌株D2对ATZ的耐受性远高于环境中的平均浓度,因此它是一株在实际污染环境中具有应用价值的降解菌株.
-
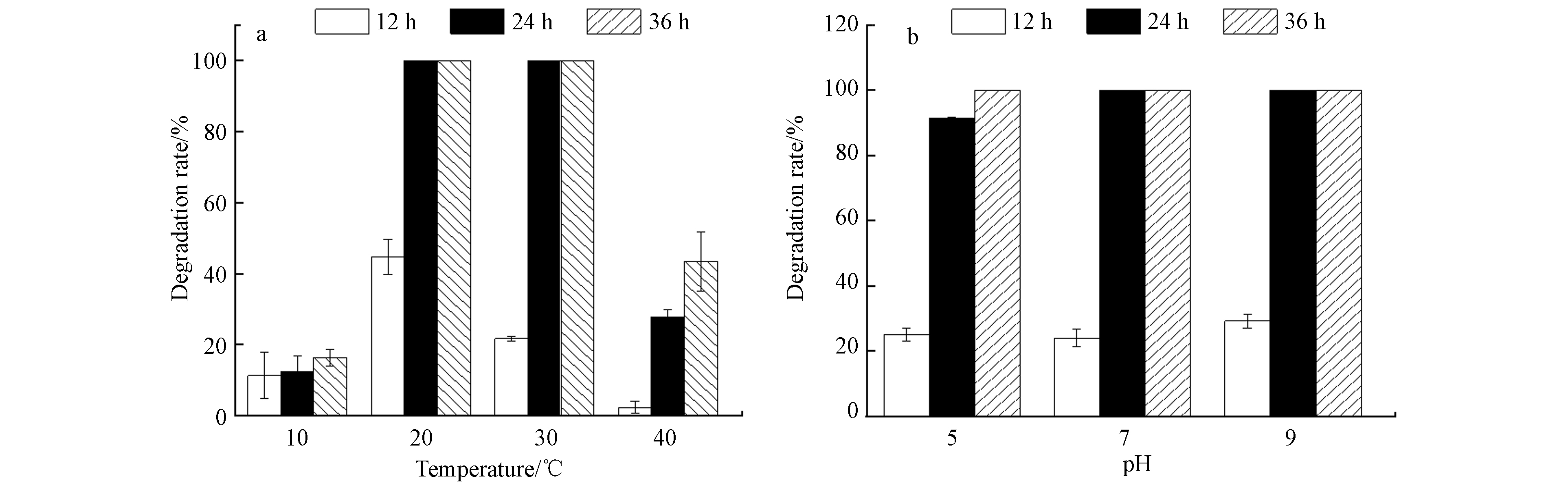
温度和pH会通过影响细菌的生长而影响其降解能力[26-28],不同温度(10、20、30、40 ℃)条件下,D2对初始浓度为100 mg·L−1 ATZ的降解情况见图5a. 当温度为20—30 ℃时,D2在24 h内将ATZ完全降解;但在10 ℃和40 ℃下培养36 h时,D2对ATZ的降解率仅为16.33%和43.48%,说明低温和高温都会抑制菌株的代谢作用,这与菌株的酶活性密切相关[29]. 另外,培养基的酸碱度也会影响细菌酶的合成和催化活性[9],细菌表面的电荷分布随着pH的改变而改变 [26]. 不同pH条件下,D2对初始浓度为100 mg·L−1(培养温度为30 ℃)ATZ的降解率如图5b所示. 当pH为5.0、7.0和9.0 时,经过24 h,D2对ATZ的降解率分别为到91.42%、100%、100%. 据以往研究报道,pH为5.0和9.0时,阿特拉津降解菌(Enterobacter sp.)的降解率低于70%[27],菌株L-6 [28] 仅能适应碱性环境,当pH=6时,降解率为45.6%,因此菌株D2对pH具有较高的适应性.
-
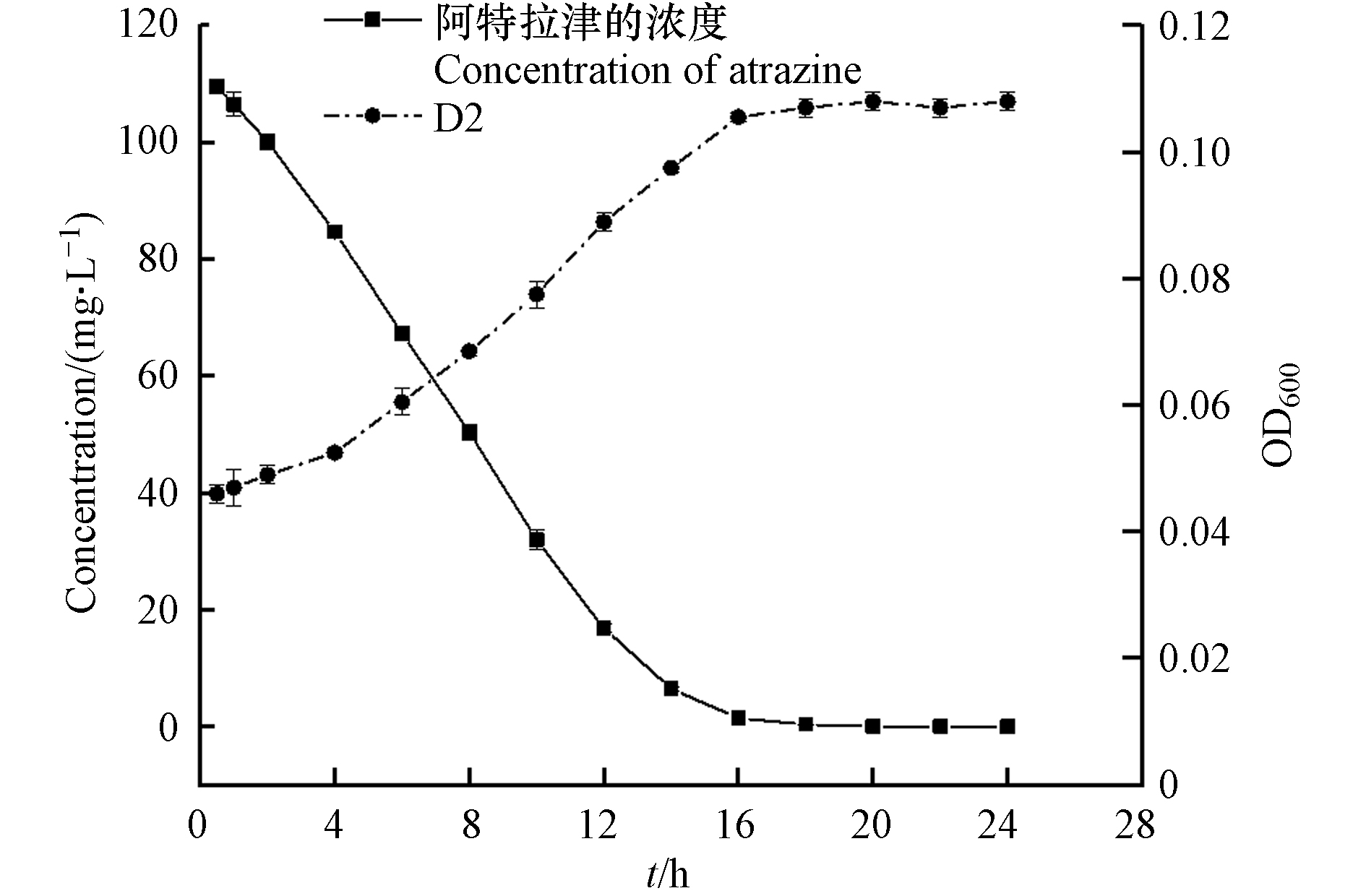
在最佳降解条件下(2%的菌悬液接种量、温度为20 ℃、pH为9.0),将菌株D2接种至初始浓度为100 mg·L−1的ATZ无机盐培养基中,生长及降解动态曲线如图6所示. 菌株D2的生长与ATZ浓度呈较强的负相关(表3,r=−0.983,P<0.01),随着菌株的生长,阿特拉津的浓度急剧降低. 培养初期D2存在短暂的适应期,OD600从0.046增至0.052;培养4 h后进入对数生长期,OD600达到0.097,此时随着菌株的大量生长ATZ被快速降解;在培养14 h以后菌株进入静止期,此时菌株数量达到最大值,其OD600为0.107,ATZ浓度降为6.41 mg·L−1,降解率达到94.33%,在18 h时ATZ被完全去除. 实验结果表明,阿特拉津浓度的降低与D2的数量密切相关,证明菌株D2具有阿特拉津去除能力.
-
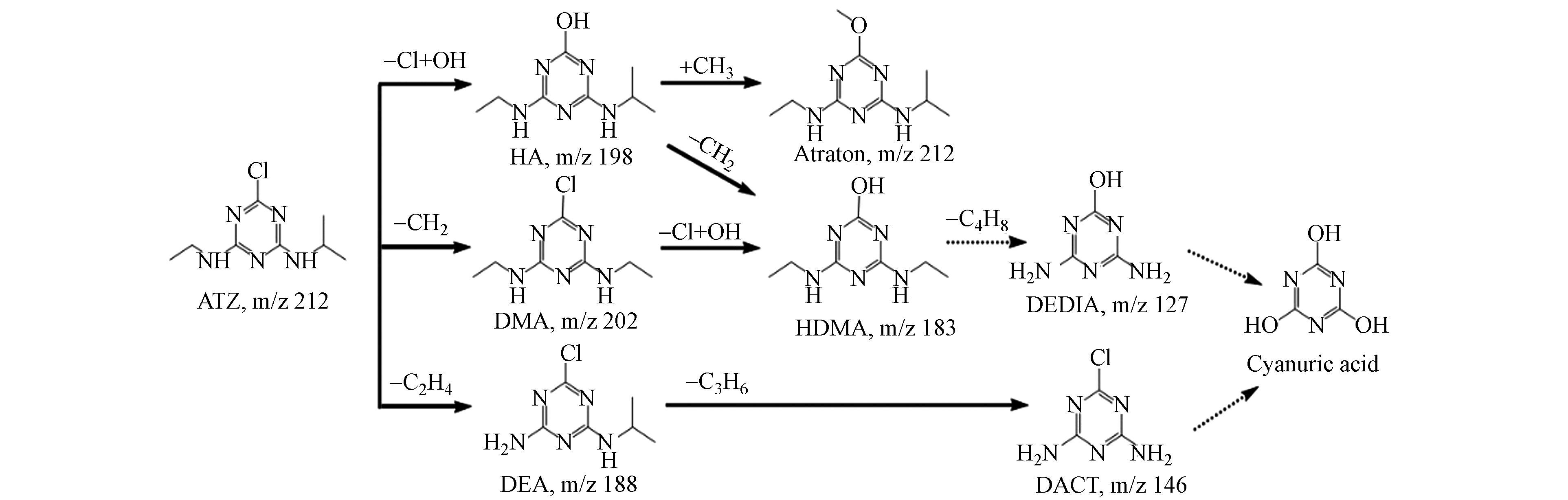
菌株D2于含ATZ的无机盐培养基进行培养,取12 h时的样品进行分析,采用高效液相色谱-质谱联用仪(HPLC-MS)测定其降解产物,共检测出6种可能的代谢物(图7 b—g),质荷比(m/z)分别为146.00、184.12、198.13、202.08、212.15、188.07,与已知标准化合物和报道的阿特拉津代谢物的比较,这6种代谢物分别被确定为脱乙基脱异丙基阿特拉津(DACT)、羟基西玛津(HDMA)、羟基阿特拉津(HA)、西玛津(DMA)、阿特拉通(Atraton)和脱乙基阿特拉津(DEA).
根据产物的结构组成和报道过的阿特拉津降解途径提出菌株D2降解阿特拉津的可能途径,结果如图8所示.
途径Ⅰ:首先羟基取代氯,形成脱氯羟基化的阿特拉津(HA),然后甲基取代HA的氢生成阿特拉通(Atraton),HA也可能通过脱甲基化反应转化为羟基西玛津(HDMA).途径Ⅱ:阿特拉津的异丙基脱甲基化生成西玛津(DMA),然后DMA的氯被一个羟基取代,转化为羟基西玛津(HDMA).途径Ⅲ:阿特拉津经N-脱烷基反应转化为脱乙基阿特拉津(DEA),随后DEA异丙基化为脱乙基脱异丙基阿特拉津(DACT). Liu等[30]在研究土壤中阿特拉津的降解途径时得到了相似的结果,阿特拉津可通过光解、水解及微生物降解转化为羟基阿特拉津、阿特拉通、扑灭津、脱乙基阿特拉津、羟基西玛津、西玛津、去乙基异丙基化阿特拉津等. 而Zhang 等[31]和郭火生等[32]还发现羟基阿特拉津可以被进一步水解为三聚氰酸.通过以上总结推测,阿特拉津的降解中间产物还会通过脱甲基化、脱烷基化和水解等产生三聚氰胺二酰胺(DEDIA)、去乙基异丙基化阿特拉津(DACT)和三聚氰酸等副产物.
-
将本研究中的D2与已报道过具有阿特拉津降解能力的菌株进行对比,结果汇总如表4所示. 目前已经筛选出很多能以阿特拉津为唯一碳源或氮源生长的菌株,并适用于阿特拉津浓度范围为8—100 mg·L−1的废水,但本研究中的新菌株D2具有更优异的降解性能. 已研究的菌株Acinetobacter lwoffii DNS32 [32]在 20 ℃和 35 ℃时,对阿特拉津的降解率约为 30%—35%,最佳生长pH为7—8,在酸性及pH高于8的条件下,菌株的生长及降解能力受到抑制;菌株C2[33]在20 ℃、30 ℃、40 ℃下培养5 d后,降解率分别为86.7%、97.6%和21.9%;菌株SB5[34]生长的温度范围是25—37 ℃,在pH=8的条件下完全降解ATZ需36 h. 本研究分离出的菌株D2在20 —30 ℃、pH为5.0 — 9.0的范围内,18 h以内即可将100 mg·L−1的阿特拉津完全降解,远远快于培养时间为24 —264 h的其他菌株,且具有更强的pH和温度的适应性,在40 ℃的高温下仍具有一定的活性,降解率高于40%,在10 ℃的条件下,D2对ATZ的降解率仍有近20%. 另外,已报道的菌株大部分来源于北部寒冷地区,而菌株D2来源于江苏省农田土壤,更能适应夏季高温、有机质及氮磷含量低、弱酸性的土壤环境,可成为长三角地区阿特拉津污染土壤和废水修复的微生物资源.
-
(1)从江苏省5个不同区域的农田土壤中筛选出一株以阿特拉津为唯一碳源和氮源生长的菌株D2,属于土壤芽孢杆菌(Solibacillus),该菌株能适应长三角地区环境,并有效降解水体和土壤中的阿特拉津.
(2)在pH 5.0—9.0、温度20—30 ℃的条件下,菌株具有良好的降解能力. 最适条件下,D2能在18 h内将100 mg·L−1的ATZ完全降解,具有较高的耐受性及降解效率.
(3)D2主要通过脱氯羟基化、加氢脱烷基化、甲基化、脱烷基化和水解等将ATZ转化为羟基阿特拉津(HA)、阿特拉通(Atraton)、脱乙基阿特拉津(DEA)、西玛津(DMA)、羟基西玛津(HDMA)和脱乙基脱异丙基阿特拉津(DACT).
一株高效阿特拉津降解菌株的筛选及其降解能力和机理
Screening and identification of an atrazine-degrading strain and its degradation capacity and mechanism on atrazine
-
摘要: 在江苏省某玉米农田土壤中筛选出以阿特拉津(ATZ)为唯一碳源和氮源的菌株D2,经16S rDNA基因序列分析,将其鉴定为土壤芽孢杆菌(Solibacillus),菌株D2的生长曲线符合SGompertz模型和Slogistic模型. 不同ATZ初始浓度、pH和培养温度条件下对ATZ的降解实验表明,菌株D2对ATZ具有极高的耐受性(>200 mg·L−1),在温度为20—30 ℃、pH值为5—9的条件下降解率均能达100%,可将100 mg·L−1的ATZ在18 h内完全去除,ATZ去除量与D2菌株的数量呈显著的负相关(r=−0.983,P<0.01). 对ATZ的降解中间产物测定表明,菌株D2可通过脱氯羟基化、加氢脱烷基化、甲基化、脱烷基化和水解等过程将阿特拉津转化为羟基阿特拉津(HA)、阿特拉通(atraton)、脱乙基阿特拉津(DEA)、西玛津(DMA)、羟基西玛津(HDMA)和脱乙基脱异丙基阿特拉津(DACT). 因此,D2是一株高效降解菌株,环境适应能力高于大部分已报道菌株,能够广泛应用于ATZ污染废水和污染土壤修复等领域.Abstract: Strain D2, using atrazine(ATZ) as the only carbon source and nitrogen source, was isolated from a maize field in Jiangsu Province. It was preliminarily identified as Solibacillus by 16S rDNA gene sequence analysis, and its growth curve both fit SGompertz model and Slogistic model. Experiments demonstrated that under different initial ATZ concentration, pH and temperature, strain D2 could tolerate high concentration of ATZ (>200 mg·L−1), wider pH and temperature, compared with reported strains. Strain D2 degraded ATZ from 100 mg·L−1 to 0 mg·L−1 within 18 h at 20—30 ℃, pH 5—9, and there was a significant negative correlation between the amount of ATZ removal and the population of D2 strain (r=−0.983, P<0.01). Analysis of the degradation intermediates of ATZ revealed that strain D2 transformed atrazine into hydroxy atrazine (HA), atraton, simazine (DMA), deethyl atrazine (DEA), hydroxy simazine (HDMA) and desethyl-desisopropyl atrazine(DACT)through dechlorination hydroxylation, hydrodealkylation, methylation, dealkylation and hydrolysis. Therefore, strain D2 is a strong degrading strain with better environmental adaptability than most reported strains, and could be used widely in the remediation of ATZ wastewater and contaminated soil.
-
Key words:
- atrazine /
- strain /
- degradation characteristics /
- degradation pathway /
- Yangtze River Delta
-
表 1 菌株D2的生理生化特性
Table 1. Physiological characteristic of strain D2
特征 Characteristic 结果 Results 革兰氏染色 + 淀粉水解 − 吲哚 − 甲基红 − V-P产生 − 柠檬酸盐利用 − 硝酸盐还原 + 过氧化氢酶 + 注:“+”为阳性;“−”为阴性. Note:“+” means positive and “−” means negative. 表 2 菌株生长曲线的SGompertz模型和Slogistic模型拟合参数对比
Table 2. Comparative list of bacteria growth due to SGompertz and Slogistic model
名称 Name SGompertz model Slogistic model R2 a1 K1 XC/h μ1/h−1 R2 a2 K2 X0/h μ2/h−1 D2 0.991 1.995 0.279 9.647 0.214 0.992 1.958 0.454 10.950 0.222 表 3 浓度与OD600的相关性分析
Table 3. Correlation analysis between concentration and OD600
平均值 Average value 标准差 Standard deviation 浓度 Concentration OD600 浓度 41.093 44.110 1 OD600 0.080 0.026 −0.983** 1 注:* P<0.05,** P<0.01. 表 4 已报道菌株对阿特拉津的降解效果的比较
Table 4. Comparison of atrazine degradation by strains has been reported
菌种 Strains 浓度/(mg·L−1)Concentation 温度/℃Temperature pH 地区Region 降解率/% Degradation 时间/h Time 文献来源Literature sources Klebsiella sp. FH1 50 25 9 吉林 81.5% 264 [13] LY-2 100 25 — 35 6 — 9 哈尔滨 98.7% 48 [17] CS3 50 30 7 河北 100% 48 [18] Arthrobacter sp. ZXY-2 50 30 — 35 8 — 9 哈尔滨 100% 6 [35] Arthrobacter sp. DNS10 100 30 7.5 哈尔滨 99.41% 24 [36] Paenarthrobacter sp. W11 100 30 7 吉林 97.1% 60 [37] Paenarthrobacter sp. W24 100 30 7 吉林 94.2% 72 [38] Arthrobacter sp. C2 100 30 7 — 9 吉林 100% 72 [33] Pseudomonas sp. 20 30 / 巴西 99% 24 [39] Achromobacter sp. 20 30 / 巴西 39% 48 [39] Pencillium sp. yz11-22N2 8 28 7 / 91.2% 120 [40] D2 100 20 — 30 5 — 9 江苏 100% 18 本研究 -
[1] ALVES de OLIVEIRA L, JARBAS HONORIO de M, GRECCO K L, et al. Atrazine movement in corn cultivated soil using HYDRUS-2D: A comparison between real and simulated data [J]. Journal of Environmental Management, 2019, 248: 109311. doi: 10.1016/j.jenvman.2019.109311 [2] CHOWDHURY I F, DORAN G S, STODART B J, et al. Trifluralin and atrazine sensitivity to selected cereal and legume crops [J]. Agronomy, 2020, 10(4): 587. doi: 10.3390/agronomy10040587 [3] HORZMANN K A, LIN L F, TASLAKJIAN B, et al. Embryonic atrazine exposure and later in life behavioral and brain transcriptomic, epigenetic, and pathological alterations in adult male zebrafish [J]. Cell Biology and Toxicology, 2021, 37(3): 421-439. doi: 10.1007/s10565-020-09548-y [4] 万年升, 顾继东, 段舜山. 阿特拉津生态毒性与生物降解的研究 [J]. 环境科学学报, 2006, 26(4): 552-560. doi: 10.3321/j.issn:0253-2468.2006.04.003 WAN N S, GU J D, DUAN S S. Eco-toxicity and biodegradation of atrazine in the environment [J]. Acta Scientiae Circumstantiae, 2006, 26(4): 552-560(in Chinese). doi: 10.3321/j.issn:0253-2468.2006.04.003
[5] CHEN Q L, YANG B S, WANG H, et al. Soil microbial community toxic response to atrazine and its residues under atrazine and lead contamination [J]. Environmental Science and Pollution Research International, 2015, 22(2): 996-1007. doi: 10.1007/s11356-014-3369-7 [6] 孙涛, 杨再磊, 蒋靖佰伦, 等. 生物炭对土壤中阿特拉津吸附特征的影响 [J]. 环境化学, 2021, 40(3): 687-695. doi: 10.7524/j.issn.0254-6108.2019102207 SUN T, YANG Z L, JIANG J, et al. Effect of biochar on the adsorption characteristics of atrazine in soil [J]. Environmental Chemistry, 2021, 40(3): 687-695(in Chinese). doi: 10.7524/j.issn.0254-6108.2019102207
[7] MAHLALELA L C, CASADO C, MARUGÁN J, et al. Photocatalytic degradation of atrazine in aqueous solution using hyperbranched polyethyleneimine templated morphologies of BiVO4 fused with Bi2O3 [J]. Journal of Environmental Chemical Engineering, 2020, 8(5): 104215. doi: 10.1016/j.jece.2020.104215 [8] MCBEATH S T, GRAHAM N J D. Simultaneous electrochemical oxidation and ferrate generation for the treatment of atrazine: A novel process for water treatment applications [J]. Journal of Hazardous Materials, 2021, 411: 125167. doi: 10.1016/j.jhazmat.2021.125167 [9] JAMES A, SINGH D K. Atrazine detoxification by intracellular crude enzyme extracts derived from epiphytic root bacteria associated with emergent hydrophytes [J]. Journal of Environmental Science and Health, Part B, 2021, 56(6): 577-586. doi: 10.1080/03601234.2021.1922043 [10] JIANG Z, JIANG D, ZHOU Q H, et al. Enhancing the atrazine tolerance of Pennisetum americanum (L. ) K. Schum by inoculating with indole-3-acetic acid producing strain Pseudomonas chlororaphis PAS18 [J]. Ecotoxicology and Environmental Safety, 2020, 202: 110854. doi: 10.1016/j.ecoenv.2020.110854 [11] GÓNGORA-ECHEVERRÍA V R, GARCÍA-ESCALANTE R, ROJAS-HERRERA R, et al. Pesticide bioremediation in liquid media using a microbial consortium and bacteria-pure strains isolated from a biomixture used in agricultural areas [J]. Ecotoxicology and Environmental Safety, 2020, 200: 110734. doi: 10.1016/j.ecoenv.2020.110734 [12] SHARMA A, KALYANI P, TRIVEDI V D, et al. Nitrogen-dependent induction of atrazine degradation pathway in Pseudomonas sp. strain AKN5 [J]. FEMS Microbiology Letters, 2019, 366(1): 277. [13] ZHANG J P, LIANG S, WANG X H, et al. Biodegradation of atrazine by the novel Klebsiella variicola strain FH-1 [J]. BioMed Research International, 2019, 2019: 4756579. [14] YANG X Y, WEI H Y, ZHU C X, et al. Biodegradation of atrazine by the novel Citricoccus sp. strain TT3 [J]. Ecotoxicology and Environmental Safety, 2018, 147: 144-150. doi: 10.1016/j.ecoenv.2017.08.046 [15] 朱希坤, 王雪涵, 姚金城, 等. Arthrobacter sp. AD30和Pseudomonas sp. AD39在阿特拉津工业废水生物处理及污染土壤生物修复中的应用 [J]. 环境化学, 2010, 29(4): 609-613. ZHU X K, WANG X H, YAO J C, et al. Biotreatment of atrazine-containing industrial wastewater and bioremediation of atrazine-contaminated soil by Arthrobacter sp. strain AD30 and Pseudomonas sp. strain AD39 [J]. Environmental Chemistry, 2010, 29(4): 609-613(in Chinese).
[16] de OLIVEIRA LOPES R, PEREIRA P M, PEREIRA A R B, et al. Atrazine, desethylatrazine (DEA) and desisopropylatrazine (DIA) degradation by Pleurotus ostreatus INCQS 40310 [J]. Biocatalysis and Biotransformation, 2020, 38(6): 415-430. doi: 10.1080/10242422.2020.1754805 [17] 李阳阳, 张金波, 沙君雪, 等. 阿特拉津降解菌LY-2的分离鉴定及其对污染土壤的修复 [J]. 农业生物技术学报, 2018, 26(6): 987-994. LI Y Y, ZHANG J B, SHA J X, et al. Isolation and identification of atrazine-degrading bacterial strain LY-2 and its bioremediation to contaminated soil [J]. Journal of Agricultural Biotechnology, 2018, 26(6): 987-994(in Chinese).
[18] 杨晓燕, 李艳苓, 魏环宇, 等. 阿特拉津降解菌CS3的分离鉴定及其降解特性的研究 [J]. 农业环境科学学报, 2018, 37(6): 1149-1158. doi: 10.11654/jaes.2017-1453 YANG X Y, LI Y L, WEI H Y, et al. Isolation, identification, and characterization of atrazine-degrading bacterial strain CS3 [J]. Journal of Agro-Environment Science, 2018, 37(6): 1149-1158(in Chinese). doi: 10.11654/jaes.2017-1453
[19] 马放, 任南琪, 杨基先. 污染控制微生物学实验[M]. 哈尔滨: 哈尔滨工业大学出版社, 2002: 1 − 280 . MA F, REN N Q, YANG J X. Contamination control microbiology experiment [M]. Harbin: Harbin Institute of Technology Press, 2002: 1 − 280(in Chinese) .
[20] 东秀珠, 蔡妙英. 常见细菌系统鉴定手册[M]. 北京: 科学出版社, 2001: 1 − 419 . DONG X Z, CAI M Y. Handbook of systematic identification of common bacteria [M]. Beijing: Science Press, 2001: 1 − 419(in Chinese).
[21] BUCHANAN R L, WHITING R C, DAMERT W C. When is simple good enough: A comparison of the Gompertz, Baranyi, and three-phase linear models for fitting bacterial growth curves [J]. Food Microbiology, 1997, 14(4): 313-326. doi: 10.1006/fmic.1997.0125 [22] 孙鹏, 孙源. 3种方法对中肋骨条藻Logistic生长模型拟合的比较研究 [J]. 江苏农业科学, 2019, 47(2): 229-232. SUN P, SUN Y. Comparative study on three methods for Logistic growth model fitting of Skeletonema costatum [J]. Jiangsu Agricultural Sciences, 2019, 47(2): 229-232(in Chinese).
[23] PASALARI H, ESRAFILI A, REZAEE A, et al. Electrochemical oxidation pretreatment for enhanced methane potential from landfill leachate in anaerobic co-digestion process: Performance, Gompertz model, and energy assessment [J]. Chemical Engineering Journal, 2021, 422: 130046. doi: 10.1016/j.cej.2021.130046 [24] 谢柄柯, 张玉, 王晓伟, 等. 菌株Desulfovibrio sp. CMX的DNRA性能和影响因素 [J]. 环境科学, 2016, 37(10): 3955-3962. XIE B K, ZHANG Y, WANG X W, et al. Performance and influencing factors of dissimilatory nitrate reduction to ammonium process by the strain Desulfovibrio sp. CMX [J]. Environmental Science, 2016, 37(10): 3955-3962(in Chinese).
[25] 陈世宇. 不同土壤中阿特拉津降解特征、降解基因分布及细菌群落演替规律[D]. 杭州: 浙江大学, 2021: 1 − 76 . CHEN S Y. Atrazine degradation characteristic, degradation genes distribution and bacterial community succession in different soils[D]. Hangzhou: Zhejiang University, 2021: 1 − 76(in Chinese) .
[26] 程吟文, 谷成刚, 王静婷, 等. 多溴联苯醚微生物降解过程与机理的研究进展 [J]. 环境化学, 2015, 34(4): 637-648. doi: 10.7524/j.issn.0254-6108.2015.04.2014031407 CHENG Y W, GU C G, WANG J T, et al. Recent advances in mechanism and processes of microbial degradation of polybrominated diphenyl ethers [J]. Environmental Chemistry, 2015, 34(4): 637-648(in Chinese). doi: 10.7524/j.issn.0254-6108.2015.04.2014031407
[27] 刘丹丹, 梁彦秋, 刘长风, 等. 莠去津高效降解菌的鉴定和降解特性分析 [J]. 农药, 2016, 55(5): 340-342,355. doi: 10.16820/j.cnki.1006-0413.2016.05.009 LIU D D, LIANG Y Q, LIU C F, et al. Identification and characterization of an atrazine degrading bacterium [J]. Agrochemicals, 2016, 55(5): 340-342,355(in Chinese). doi: 10.16820/j.cnki.1006-0413.2016.05.009
[28] 李绍峰, 朱静, 李铁晶. 阿特拉津降解菌株的分离、鉴定及降解特性研究 [J]. 环境科学, 2012, 33(9): 3214-3219. LI S F, ZHU J, LI T J. Isolation, identification and characterization of an atrazine degrading bacterium [J]. Environmental Science, 2012, 33(9): 3214-3219(in Chinese).
[29] 赵婉琳, 叶静, 张娜, 等. 褐藻胶降解菌的筛选、鉴定及产酶条件优化 [J]. 微生物学报, 2019, 59(1): 169-180. ZHAO W L, YE J, ZHANG N, et al. Screening, identification and fermentation optimization of an alginate-degrading strain [J]. Acta Microbiologica Sinica, 2019, 59(1): 169-180(in Chinese).
[30] LIU J T, ZHOU J H, GUO Q N, et al. Physiochemical assessment of environmental behaviors of herbicide atrazine in soils associated with its degradation and bioavailability to weeds [J]. Chemosphere, 2021, 262: 127830. doi: 10.1016/j.chemosphere.2020.127830 [31] ZHANG Y, JIANG Z, CAO B, et al. Metabolic ability and gene characteristics of Arthrobacter sp. strain DNS10, the sole atrazine-degrading strain in a consortium isolated from black soil [J]. International Biodeterioration & Biodegradation, 2011, 65(8): 1140-1144. [32] 郭火生, 王志刚, 孟冬芳, 等. 阿特拉津降解菌株DNS32的降解特性及分类鉴定与降解途径研究 [J]. 微生物学通报, 2012, 39(9): 1234-1241. GUO H S, WANG Z G, MENG D F, et al. Degradation characteristics and identification and the degradation pathway of the atrazine-degrading strain DNS32 [J]. Microbiology China, 2012, 39(9): 1234-1241(in Chinese).
[33] CAO D T, HE S H, LI X, et al. Characterization, genome functional analysis, and detoxification of atrazine by Arthrobacter sp. C2 [J]. Chemosphere, 2021, 264: 128514. doi: 10.1016/j.chemosphere.2020.128514 [34] 陆长鸣, 李想, 徐明恺, 等. 一株高效广谱莠去津降解菌SB5的生长和降解特性[J]. 应用生态学报, 2022, 33 (1): 229-238 . LU C M, LI X, XU M K, et al. Growth and degradation characteristics of an efficient and broad-spectrum atrazine-degrading strainSB5[J]. Chinese Journal of Applied Ecology, 2022, 33 (1): 229-238(in Chinese) .
[35] ZHAO X Y, WANG L, MA F, et al. Characterisation of an efficient atrazine-degrading bacterium, Arthrobacter sp. ZXY-2: An attempt to lay the foundation for potential bioaugmentation applications [J]. Biotechnology for Biofuels, 2018, 11: 113-123. doi: 10.1186/s13068-018-1113-0 [36] 张庆媛, 葛世杰, 姜昭, 等. 高效阿特拉津降解菌株DNS10降解条件优化 [J]. 环境工程学报, 2013, 7(3): 1169-1174. ZHANG Q Y, GE S J, JIANG Z, et al. Optimization for degradation conditions of atrazine-degrading strain DNS10 [J]. Chinese Journal of Environmental Engineering, 2013, 7(3): 1169-1174(in Chinese).
[37] CHEN S M, LI Y Y, FAN Z W, et al. Soil bacterial community dynamics following bioaugmentation with Paenarthrobacter sp. W11 in atrazine-contaminated soil [J]. Chemosphere, 2021, 282: 130976. doi: 10.1016/j.chemosphere.2021.130976 [38] 范作伟, 李阳阳, 陈帅民, 等. 阿特拉津降解菌W24的分离鉴定和土壤修复效果研究 [J]. 玉米科学, 2021, 29(5): 172-177. FAN Z W, LI Y Y, CHEN S M, et al. Isolation and identification of atrazine-degrading bacterial strain W24 and its remediation effect on soil [J]. Journal of Maize Sciences, 2021, 29(5): 172-177(in Chinese).
[39] TONELLI FERNANDES A F, BRAZ V S, BAUERMEISTER A, et al. Degradation of atrazine by Pseudomonas sp. and Achromobacter sp. isolated from Brazilian agricultural soil [J]. International Biodeterioration & Biodegradation, 2018, 130: 17-22. [40] YU J P, HE H J, YANG W L, et al. Magnetic bionanoparticles of Penicillium sp. yz11-22N2 doped with Fe3O4 and encapsulated within PVA-SA gel beads for atrazine removal [J]. Bioresource Technology, 2018, 260: 196-203. doi: 10.1016/j.biortech.2018.03.103 -







 下载:
下载: