-
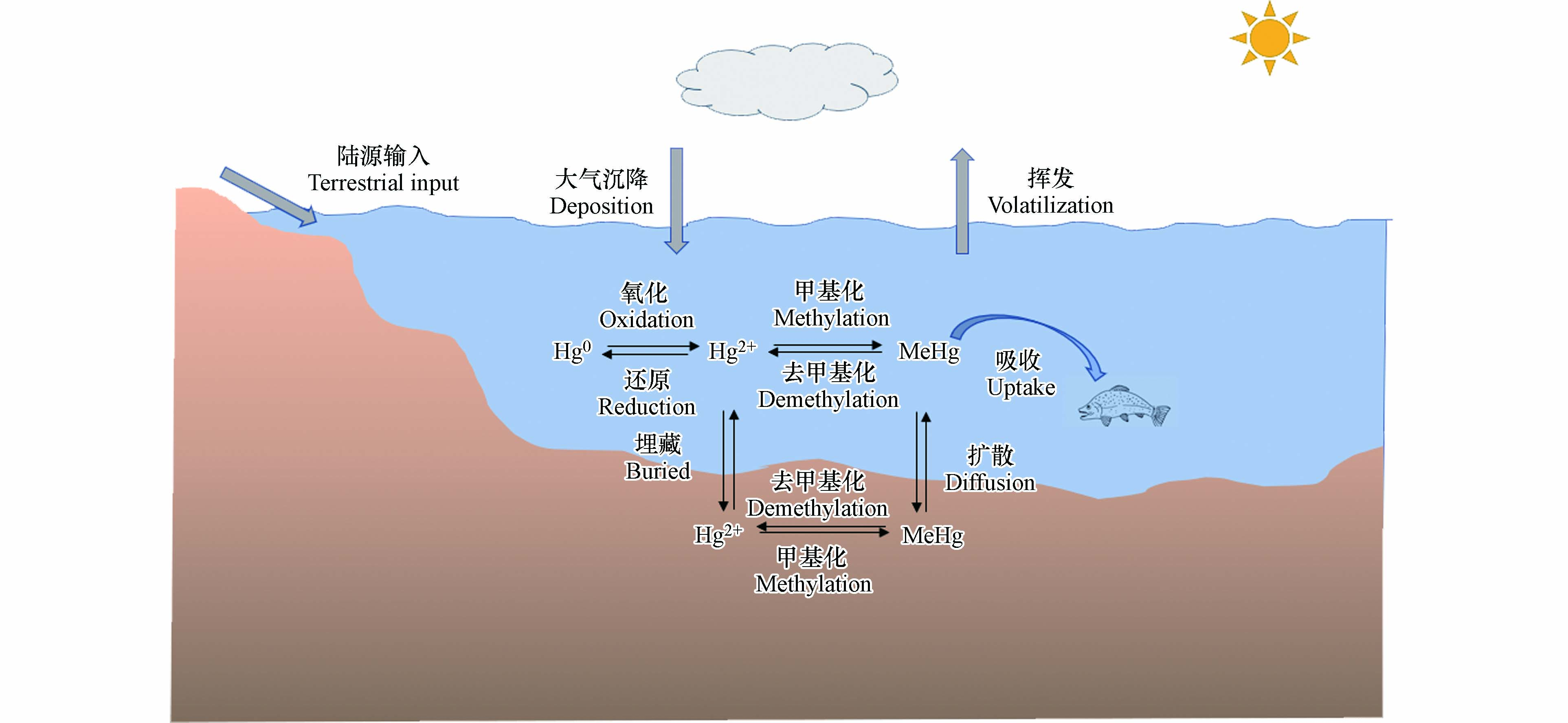
汞(Hg)是一种在大气中具有较长停留时间并能进行长距离传输的全球性有毒重金属[1-4]. 汞具有高挥发性、生物累积性和持久性的特点,对人类健康和环境造成严重危害[5-6]. 汞可通过自然过程(如岩石风化、地热活动和火山爆发)和人类活动(如采矿、工业生产和化石燃料燃烧)排放进入大气[7-9]. 海洋在全球汞循环中发挥着重要作用[10]. 2018年联合国环境规划署报告指出,每年排放到大气中的汞约有3700 t会沉降到海洋[11]. 自工业革命以来,随着人为排放汞的增加,海洋中汞浓度显著增加[12],导致海洋鱼类体内汞浓度升高,对人类健康造成巨大威胁[13-14]. 海洋环境中不同形态的汞在迁移转化过程中始终处于一个动态平衡状态,主要控制过程包括甲基化/去甲基化、吸附/解吸、沉积物-水界面交换、氧化/还原等(图1). 其中,吸附/解吸、沉积物-水界面交换等迁移过程控制着汞在不同介质中的分配,氧化/还原过程控制着水体中Hg0的水平,而甲基化/去甲基化是控制海洋环境中甲基汞循环的关键过程.
汞在环境中的主要赋存形态包括:Hg0、Hg2+和MeHg[15-19]. MeHg是一种毒性强、易富集、具有严重生态风险的有毒污染物[5, 20-22],这种生物可利用性高的汞形态能够在高营养级生物体内累积到较高浓度水平[20-21, 23]. 食用海产品是人类暴露MeHg的主要途径,可以导致神经发育障碍和自身免疫反应等疾病[20-21, 24-25]. 通过各种来源排放到海洋环境中的无机汞在沉积物和水柱中均可甲基化为MeHg. 近海沉积物中产生的MeHg也可通过长距离传输进入大洋. 原位生成是海洋甲基汞的主要来源,原位甲基化和去甲基化被认为是控制海洋环境中甲基汞水平的重要过程[18-19, 26-27]. 汞的甲基化和去甲基化均可以通过生物和化学(光化学和非光化学)途径发生[28-29],微生物途径被认为是海洋生态系统中甲基汞生成降解的主要途径[30-33]. 光化学途径在表层海水汞迁移转化过程中发挥着重要作用[34-35].
目前已有综述对环境汞的甲基化与去甲基化过程进行了总结[36-39],但对海洋环境中汞甲基化/去甲基化涉及相对较少. 本文在总结海洋汞甲基化/去甲基化途径、过程机制和微生物类群等基础上,详细讨论了海洋微生物(细菌、古菌和微藻)、海洋光化学以及海洋非光化学汞甲基化/去甲基化过程机制.
-
稳定同位素添加技术具有较高的准确度、精密度和可同时测定甲基化(km)和去甲基化速率(kd)的优点,是测定汞甲基化/去甲基化速率的有效工具[40-41]. 近年来,该技术已被广泛应用于MeHg净产量的估算,以解决水生系统中MeHg的原位生成和控制因素等问题[42-43]. 表1总结了目前报道的稳定同位素添加技术测得的海洋体系中沉积物和水体汞甲基化与去甲基化速率,其中沉积物km范围为0.001—0.21 d−1,kd为0.02—6.4 d−1;水体km范围为0.001—0.005 d−1,kd为0.09—0.57 d−1. 大洋与近岸水体比较,近岸水体汞甲基化与去甲基化速率较高;海洋体系与淡水体系比较发现,海洋体系沉积物中汞甲基化和去甲基化速率更高.
-
沉积物被认为是产生MeHg最重要的场所[57]. 一项在北大西洋的研究表明,硫循环驱动了从近海到深海峡谷沉积物汞的甲基化过程,硫酸盐还原菌(sulfate-reducing bacteria,SRB)是主要的汞甲基化微生物[58]. 南海沉积物汞甲基化过程实验结果证实,微生物甲基化是该研究区域汞甲基化的重要途径,硫酸盐还原菌和铁还原菌(iron- reducing bacteria,FeRB)是主要的甲基化微生物[59]. 汞去甲基化也广泛地发生在墨西哥湾北部、长岛海峡、大西洋中部大陆架和斜坡等海洋沉积物环境中[44-47]. 由此可见,海洋沉积物环境是汞甲基化/去甲基化的重要热点区域.
除沉积物外,汞甲基化/去甲基化过程也可能发生在海洋系统的水柱中. 最近研究表明,海洋次表层可能是汞甲基化的一个潜在热点区域. 例如,研究发现北冰洋大部分MeHg净生产发生在次表层[60]. 萨尼奇湾汞甲基化功能基因hgcAB在200 m深度的样品中丰度最高,表明潜在的新型海洋汞甲基化微生物可能具有更强的耐氧能力和更广的栖息范围[30]. 开阔大洋汞甲基化过程的研究提出,含氧水柱中的好氧微生物可能是次表层汞甲基化的主要微生物[61-64]. 与之相反,在赤道北太平洋次表层溶解氧最小区的研究发现,厌氧产甲烷菌是导致水柱中甲基汞浓度升高的主要微生物[65]. 海洋生物汞同位素组成测定结果也表明汞甲基化过程可能主要发生在浅层海水,即50—400 m深度之间[31, 66]. 太平洋中部汞甲基化和去甲基化的研究揭示,非生物甲基化/去甲基化过程在控制寡营养区域次表层水体甲基汞浓度方面发挥着重要作用[52]. 海底热液系统可能释放Hg和MeHg,因此也被认为是海洋中MeHg的潜在重要来源. 例如,戈尔达海岭(Gorda Ridge)的热液喷口流体中存在高比例的MeHg,研究者认为这可能是由系统内汞的非生物甲基化产生[67]. 微生物宏基因组结果也表明海底热液存在具有汞甲基化潜能的嗜热古菌Pyrococcus furiosus,可能是潜在的甲基化热点区域[64]. 热液喷口和地热温泉等高温环境中微生物去甲基化过程也较为活跃. 微生物去甲基化最早的分支来自嗜热细菌Aquificales,与该菌的栖息地——热液环境中高浓度的Hg2+一致[68]. 随着宏基因组学和单细胞基因组学等测序技术在海洋环境中的广泛应用,已有6个门的去甲基化微生物在热液和地热泉等环境中被鉴定出来[69].
-
人们普遍认为汞甲基化主要由微生物介导,但如果存在合适的甲基供体,无机汞也可以通过非生物途径发生甲基化反应[33, 70]. 根据是否需要光照,非生物汞甲基化分为光化学和非光化学甲基化[18]. 非生物汞甲基化过程作为汞在氧化海水中甲基化的可能途径,对海洋水体MeHg的生成也有一定贡献[27, 52-53]. 太平洋中部的一项研究发现,相对于未过滤海水,过滤海水中汞甲基化速率更高[52]. 由于海水中大部分微生物已通过0.2 μm滤膜去除掉,因此过滤海水中汞甲基化能力增强最有可能归因于非生物机制,而不是由微生物汞甲基化所引起[52]. 此外,同位素示踪实验研究发现,加入同位素标记的无机汞后会立即发生汞的甲基化,这种现象只能通过非生物汞甲基化来解释[27, 52-53, 71-72]. 非生物汞甲基化在海洋环境中的作用也得到最近一项模拟全球海洋汞甲基化过程研究的支持。该研究表明,如果不考虑非生物汞甲基化,模拟的北冰洋和南大洋甲基汞浓度将被大大低估[73].
-
研究表明,可能促进海洋汞非生物甲基化的化合物包括碘甲烷(CH3I)和二甲基硫(DMS)等. CH3I在海洋环境中由蓝藻等藻类或真菌产生,而DMS主要由海洋浮游植物产生[70, 74]. 有研究表明CH3I诱导的光化学汞甲基化可以在水生环境发生,但该研究设置的CH3I的浓度(mmol·L−1水平)远高于海水(pmol·L−1水平)中的实际浓度,该过程在海水中的贡献尚有待验证[70, 75]. 此外,也有研究表明溶解有机质(DOM)驱动的光化学汞甲基化在降水和湖泊MeHg生成中起着重要作用,但其在海洋环境中的作用尚不清楚[76-77]. 目前普遍认为光化学汞甲基化不太可能成为海洋中甲基汞的主要来源,海洋表层水体MeHg浓度总体较低也支持了这一观点[19].
-
生物去甲基化在海水(尤其是深层水体)和沉积物MeHg降解中起着重要作用. 非生物去甲基化也可发生,且大部分研究集中在光化学去甲基化上,这一过程对海洋MeHg降解的贡献可以达到56%—80%[78-79]. MeHg可以通过直接或间接的光化学反应进行降解. 直接光降解利用海水中MeHg的C—Hg键或MeHg-DOM络合物的Hg—S键吸收光,导致键的直接能量转移和断裂[14, 80]. 间接光降解一般归因于光化学诱导产生的活性氧和自由基(如过氧化氢、羟基自由基、单线态氧、过氧化物和DOM的激发三重态)对MeHg的降解[79, 81-82]. 光强、光波长以及水体的理化参数等环境因素均对海水甲基汞光降解产生影响.
赤道太平洋、北冰洋和白令海峡等海域的MeHg光化学去甲基化现场培养实验表明,MeHg降解速率与光强度和波长之间存在明显的相关性[26]. 一般认为,紫外线(UV)是导致MeHg光降解的主要光波段,波长范围为280 nm至400 nm的UV-A和UV-B可以显著影响MeHg的光降解过程[79, 81, 83-84]. 利用汞同位素的非质量分馏(Mass-independent fractionation,MIF)特征示踪海洋生物MeHg的来源时发现,MIF与太阳辐射能量之间密切相关,光化学去甲基化是太平洋表层水中甲基汞降解的重要途径[66, 85]. 除光谱影响外,海水中的DOM也可以通过多种方式影响MeHg的光降解速率。例如,通过形成MeHg-DOM络合物或光化学反应产生自由基促进MeHg去甲基化,或者通过淬灭自由基或吸收太阳辐射降低光化学去甲基化速率[14, 79-80, 86].
-
如图1所述,海洋环境中MeHg的水平受到甲基化和去甲基化过程的共同控制[42, 87],其中微生物过程是甲基汞生成降解的最主要途径[33, 87-88].
-
微生物汞甲基化可以将海洋环境中的无机汞转化为甲基汞,了解微生物汞甲基化的生化机制至关重要[63-64]. 在发现hgcAB基因对汞甲基化的重要作用后,乙酰辅酶A途径被认为是微生物汞甲基化的主要生化机制[89]. 基于SRB和FeRB两种模型菌株,研究者提出hgcA基因能够编码一种起催化甲基转移作用的类咕啉蛋白(HgcA),而hgcB基因会编码一个与HgcA相关的铁氧化还原蛋白(HgcB),两种蛋白能将Hg通过酶催化的甲基化反应生成MeHg[89-90]. hgcAB基因是汞甲基化能力的预测因子,hgcAB基因的发现有助于人们在不同海洋环境中寻找潜在的汞甲基化微生物[57, 64]. 迄今为止,经实验培养验证的海洋环境中汞甲基化微生物主要来自δ-变形菌门(SRB、FeRB和互营养菌)、厚壁菌门和古菌门(产甲烷菌)的厌氧菌[91]. 海洋环境中已确定为汞甲基化微生物或具有 hgcAB同源物的微生物如表 2 所示。
目前在南大洋、大西洋、西太平洋、赤道太平洋、东海和里海等许多海洋环境的沉积物和水体中均检测到hgcAB基因和汞甲基化微生物的存在[64, 92]. 萨尼奇湾的研究发现,携带hgcAB基因的δ-变形菌在200 m深度的海水中具有较高的丰度[30]. 在南海获得的沉积物样品中检测到的汞甲基化细菌属于3个微生物门,包括δ-变形菌门、厚壁菌门和广古菌门,其中δ-变形菌所携带的hgcAB基因丰度最高,是最主要的汞甲基化微生物[59]. 北冰洋宏基因组中检测到具有hgcAB基因的SRB脱硫杆菌科的序列,赤道北太平洋宏基因组中则鉴定出来自产甲烷菌所携带的hgcAB基因[65]. 在波罗的海的缺氧水体和海洋雪中也发现hgcAB基因,系统发育分析表明缺氧水中δ-变形菌是最主要的汞甲基化微生物[93]. 虽然目前已鉴定的汞甲基化菌株多为厌氧菌,但好氧微生物对海洋汞甲基化过程的贡献也不能忽视[64]. 例如,携带hgcAB基因的海洋微嗜氧硝化刺菌((Nitrospina)被认为是南极海冰、西北太平洋和北冰洋等海洋环境中潜在的汞甲基化微生物[61-63, 65].
微生物汞甲基化是一个复杂的过程,可能会受到温度、pH、氧化还原电位(Eh)、DOM等各种生物地球化学因素的影响[99]. 温度升高通常会刺激微生物活动,导致汞甲基化速率增加. 在北极沿海沉积物的研究中发现温度升高会导致SRB活性增强,甲基化速率也随之增加[100]. 研究报道低pH值可以促进甲基汞的生成:一方面是因为Hg2+生物可利用度随着pH值的降低而增加;另一方面是因为汞甲基化微生物在较低pH值下能占据主导地位,从而促进甲基化过程的发生[101]. 一项在波罗的海的研究表明,海洋沉积物中微生物甲基化速率与Eh存在负相关关系[102]. 在影响汞甲基化速率的所有生物地球化学因素中,DOM被认为是最重要的因素之一[103]. DOM可以通过两种方式影响汞的生物可利用性和甲基化潜力[99]:一方面,Hg与DOM形成的络合物难以通过细胞膜扩散,从而降低汞的生物可利用性[104];另一方面,许多研究发现沉积物中MeHg浓度与DOM含量具有较好的正相关关系,这可能是由于DOM作为生长所需的营养物质促进了汞甲基化微生物的生长[105].
-
微生物去甲基化是MeHg在海洋水体和沉积物中进行降解的主要途径[23, 106-107]. 绝大多数被鉴定为能发生汞甲基化过程的微生物也具有降解MeHg的能力[96, 108]. 因此,在总体评估海洋环境中汞甲基化潜力时,应同时考虑微生物对MeHg的去甲基化作用.
-
研究表明微生物降解甲基汞具有两种途径:第一种为mer操纵子系统介导的还原去甲基化,导致Hg0和CH4的生成[107, 109-114]; 第二种为氧化去甲基化,MeHg被降解为Hg2+、CO2和少量CH4[18, 23, 108]. mer体系广泛分布于原核生物中,利用Hg0的低水溶性和易挥发的特点,原核生物可以将Hg2+还原为Hg0,使其能够在高汞浓度下生长[106, 115]. 该系统由汞还原酶(MerA)和多种运输功能蛋白质组成,并受到MerR的精细调控[106, 115]. MerR既是功能性基因表达的抑制因子,也是功能性基因表达的激活因子[106-107, 109-111, 114]. 编码这些不同功能的基因集中在主操纵子中[106-107, 110]. 少数抗汞细菌,即所谓的广谱抗汞细菌,除了对Hg2+有抗性外,还对有机汞化合物有抗性,并编码有机汞裂解酶(MerB)[112-113, 116-119].
参与MeHg还原去甲基化的微生物所拥有的mer操纵子主要位于质粒、染色体、转座子和整合素中,这意味着mer操纵子可以很容易地转移到其他微生物中,因此mer操纵子分布广泛[107]. 在南极海冰、红海的深海和热盐水环境中均检测到merA基因的存在[69],merB基因也在赤道附近的北太平洋和北冰洋水域被检测到[30, 65, 120-121]. 研究发现赤道北太平洋溶解氧最小区mer基因的相对丰度超过了hgcA基因的相对丰度,与该区域水层观测到的MeHg净去甲基化现象一致[65]. 在萨尼奇海湾,merB基因的相对丰度在200 m深处达到峰值,约占微生物群落总基因的0.6%,同时也观察到随merB基因丰度的增加MeHg水平下降的现象[30]. 目前,已在不同海域分离鉴定出290种具有还原去甲基化能力的海洋异养细菌,其中244种被归类为Alteromonas属,46种是Marinobacter属[122].
-
与mer操纵子介导的还原去甲基化过程相比,氧化去甲基化过程的研究相对较少[106]. 氧化去甲基化是一个非特异性的代谢过程,与汞的解毒无关,在缺氧环境中常见[106-107, 123-126]. 虽然缺乏纯培养体系对氧化去甲基化过程的验证,但SRB和产甲烷菌仍被认为是其主要参与者[106-108, 114]. 氧化去甲基化过程之所以被如此命名,是因为去甲基化的最终产物是氧化形式的Hg2+、CO2和少量的CH4[23, 108, 124-125]. SRB和产甲烷菌参与氧化去甲基化的区别在于前者可以产生H2S和CO2(方程1),而后者产生CH4和CO2(方程2)[23, 106, 124, 126]. 对于这种现象,研究者推测可能与SRB和产甲烷菌的一碳(C1)代谢有关[23, 106, 108, 124, 126]. 与携带MerB的微生物的活性解毒机制不同,氧化去甲基化是微生物代谢的一部分,甲基汞可能作为电子供体产生作用[87, 106-107, 126]. 据推测,SRB降解MeHg的机制与乙酸盐氧化的机制相似(方程1),而产甲烷菌对MeHg的共同代谢则通过类似于一甲胺降解的途径进行(方程2)[106-107, 115]:
虽然这些假设的氧化去甲基化途径是合理的,但尚未得到实验验证. 海洋和沉积物中氧化降解甲基汞的细菌菌株尚未被分离或鉴定,氧化去甲基化的途径仍有待深入研究[106, 108, 114].
-
海洋微藻是海洋生态系统中最重要的初级生产者,其对汞的吸收和分子转化是海洋中汞生物积累的重要过程[127-128]. 目前仅有少数研究探究了海洋微藻对汞的甲基化和去甲基化作用,且不同研究的结果存在矛盾[127, 129-130]. 早期研究发现海洋硅藻三角褐指藻能在高浓度HgCl2 (20—120 μg·L−1)条件下诱导Hg的甲基化,作者们认为该过程发生在藻细胞内部,并向外界环境释放MeHg[129]. 但由于实验中添加的汞含量远高于其在天然海水中的浓度,对于海洋微藻是否参与自然海洋环境中汞的甲基化过程仍然缺乏明确的认识. 一项利用同位素示踪技术测定绿藻汞甲基化能力的研究表明,在模拟自然海水条件下莱茵衣藻并不能导致汞的甲基化[131]. 最近我们测试了4门15种海洋微藻的汞甲基化能力,发现被测微藻均未显示出对无机汞的甲基化能力,表明浮游植物在海洋环境甲基汞生成中的作用可能可以忽略不计[132]. 三项研究结果的差异可能是由于高浓度Hg2+条件下,微藻分泌的由脂肪酸和多肽等物质组成的DOM会引起汞的化学甲基化反应[132-134].
相较于机制不明的甲基化过程,有研究发现藻细胞介导的可见光对MeHg的去甲基化可能占水体光化学驱动的去甲基化总量的20%—55%[130],在甲基汞降解中起着重要作用. 放射性同位素(CH3203Hg)实验结果表明,MeHg可以被球石藻降解为Hg0[127]. 同位素示踪研究发现莱茵衣藻能导致MeHg降解,藻细胞内合成的谷胱甘肽以及植物螯合肽可以极大地促进MeHg的去甲基化过程[129]. 最近我们测试了15种常见海洋微藻对汞的去甲基化能力,发现微藻胞外分泌物引起的MeHg光去甲基化速率在0.01—0.39 d−1之间,且部分微藻的共生菌也能诱导MeHg的去甲基化(0.03—0.14 d−1)[132]. 微藻胞外分泌物介导的MeHg光去甲基化速率与现场MeHg光去甲基化和微生物去甲基化速率相当,表明微藻在海洋环境MeHg的去甲基化中发挥着重要作用. 总结可知,藻类介导的MeHg的去甲基化可能主要是由其胞外分泌物(通过光介导途径)引起的,而不是微藻细胞对MeHg的直接去甲基化[127, 131-132]. 进一步研究也发现巯基可能是微藻胞外分泌物中导致MeHg光去甲基化的主要成分[132, 135].
-
非光化学Hg甲基化包括以有机金属配合物作为甲基供体的转甲基化[136],以甲基钴胺素为甲基供体的细胞外汞甲基化[137],以及腐殖质对汞的甲基化[138]. 目前已经证明在海洋环境中除了甲基锡之外,其余各种有机金属化合物的转甲基化途径都无关紧要[70, 139]. 然而,Celo等研究使用的甲基锡浓度远高于海水中甲基锡的测量值,因此甲基锡化合物介导的汞非生物甲基化可能也并不是海水中甲基汞的主要来源[70]. 细胞外汞甲基化主要指由生物体释放到环境中的甲基钴胺素导致的非酶化汞甲基化,由此将其与hgcAB介导的细胞内汞甲基化(也涉及甲基钴胺素)区分开来[89]. 研究发现在甲基钴胺素介导的细胞外汞甲基化过程中,Hg2+作为亲电试剂攻击甲基钴胺素,导致解离出CH3−,随后CH3−转移到高氧化态的Hg2+上生成MeHg[140-141]. 对于腐殖质介导的汞化学甲基化,CH3−向Hg2+的转移被认为是主要的甲基化途径[134]. 由于腐殖质的组成不同,汞甲基化所涉及的化合物组分不同,反应动力学也存在差异,并且对腐殖质中特定甲基供体的汞甲基化缺乏了解,因此尚无法定量估计其对海水中MeHg的贡献[138, 142]. 总体而言,虽然很难估计非生物汞甲基化对海水MeHg的贡献,但已有的研究表明,甲基钴胺素和腐殖质介导的汞甲基化在海水中可能起着一定作用[52, 73].
-
虽然大多数关于MeHg降解的研究主要集中在光化学和微生物去甲基化,但非光化学去甲基化也已被证实可以在水环境中发生. 目前有两种化学去甲基化途径已在研究中得到证实:(1)是甲基汞与H2S或硫化物矿物反应生成HgS(s)和二甲基汞(DMeHg)[143-145];(2)是MeHg与硒氨基酸反应生成HgSe(s)[146-147].
最近的研究证明,MeHg吸附在硫化物矿物或有机二硫醇表面会形成DMeHg[143-144]. 硫化物矿物上多个S原子与Hg2+进行配位形成稳定的过渡态,并激活C—Hg键以进行甲基转移或形成DMeHg[143, 148-149]. MeHg和H2S之间的反应通过强的Hg—Hg相互作用激活甲基在分子内转移,诱导(CH3Hg)2S复合体的重排或甲基直接从一个取代基转移到另一个取代基,从而促进形成DMeHg[148]. 这些重要的发现表明矿物表面的硫基减少可以导致MeHg的降解,并伴随海洋环境中DMeHg的生成[143-144]. 与硫化物介导的去甲基化途径类似,硒介导的去甲基化被认为涉及MeHg和硒氨基酸之间的反应,通过形成(CH3Hg)2Se和DMeHg作为中间体,最终降解产物为HgSe(s)[146-147]. 虽然这些反应是在相对较高的硒水平下观察到的,但这种DMeHg去甲基化的机制已经在海洋哺乳动物的肝脏和肠道中被检测到[150-151]. 这些结果表明,硒可以将MeHg转化为无机汞,从而降低其生物可利用性和生物积累.
-
(1)不同汞甲基化/去甲基化途径贡献估算:汞甲基化/去甲基化包括微生物、光化学和非光化学等多种途径,量化各途径贡献是认识MeHg迁移转化循环的基础,改进汞同位素分馏技术并结合同位素添加示踪技术有望实现区分非生物与生物甲基化/去甲基化相对贡献的目标.
(2)实际海洋环境汞甲基化/去甲基化基因/微生物作用验证:虽然目前已有一些关于海洋环境中汞甲基化/去甲基化微生物和功能基因的初步报道,但对这一过程中关键微生物和功能基因分布及其在海洋汞循环过程中的作用仍缺乏基本了解, 解决这一问题将有助于理解hgcAB和merAB等功能基因及相关微生物在实际海洋环境中的作用,并为预测海洋系统中汞的长期风险提供数据支撑.
(3)定量环境因素对海洋汞甲基化/去甲基化的影响:海洋汞甲基化/去甲基化过程受到多种物理、化学与生物因素的影响,包括温度、Eh、pH、无机汞的形态和浓度、DOM和微生物群落结构等. 定量这些环境因素对海洋汞甲基化/去甲基化的影响是预测未来气候变化和生态系统演变背景下汞长期风险的关键.
海洋环境汞甲基化/去甲基化研究进展
Progress in mercury methylation and demethylation in marine environment
-
摘要: 汞(Hg)是一种在大气中具有较长停留时间并能进行长距离传输的全球性污染物. 甲基汞(MeHg)具有毒性强、易富集、可随食物链放大的特点,是引起环境风险的主要汞形态. 通过各种来源排放到海洋环境中的无机汞在沉积物和水柱中均可甲基化为MeHg,原位甲基化和去甲基化是控制海洋环境中甲基汞水平的关键过程. 虽然目前已有综述总结了环境中汞甲基化/去甲基化,但对海洋生态系统中汞甲基化/去甲基化过程涉及相对较少. 本文在总结海洋汞甲基化/去甲基化过程速率、途径和热点区域基础上,详细讨论了海洋光化学、非光化学以及微生物3种汞甲基化/去甲基化途径机制,并对未来研究方向进行了展望. 在现有研究基础上,未来应在不同汞甲基化/去甲基化途径贡献估算、实际海洋环境汞甲基化/去甲基化基因/微生物作用验证、环境因素对海洋汞甲基化/去甲基化影响方面开展更深入研究. 海洋汞甲基化/去甲基化的研究可为深入理解海洋汞的环境行为与风险和发展有效的汞污染风险防控技术提供科学依据和数据支撑.Abstract: Mercury (Hg) is a global pollutant since it has a long residence time in the atmosphere and can be transported over a long distance. Methylmercury (MeHg) has the characteristics of high toxicity, and bioaccumulation along the food chain. It is the major form of Hg that causes environmental risks. Inorganic Hg discharged into the marine environment through various sources can be methylated into MeHg in sediment and water column. In situ methylation and demethylation are the key processes that control the level of MeHg in marine environments. Although the methylation and demethylation of Hg in the environment have been reviewed in several papers, both processes in the ocean have not been comprehensively reviewed yet. In this review, the rates, pathways and hot spots of Hg methylation/demethylation in marine environments were summarized first. Then, the mechanisms of the three Hg methylation/demethylation pathways in marine environments, i.e., photochemical pathway, non-photochemical pathway, and microbial pathway were discussed in detail. Future perspectives of this research area were also discussed. On the basis of existing research, more in-depth research should be carried out in the future on the contribution of different Hg methylation/demethylation pathways, the verification of the importance of Hg methylation/demethylation genes/microbes in Hg methylation/demethylation in natural marine environments, and the impact of environmental factors on marine Hg methylation/demethylation. The research on methylation/demethylation of Hg in the ocean can provide scientific basis and data support for in-depth understanding of environmental behaviors and risks of mercury in marine environments and the development of effective prevention and control techniques for Hg.
-
Key words:
- marine /
- mercury /
- methylation /
- demethylation /
- mechanism.
-
汞(Hg)是一种在大气中具有较长停留时间并能进行长距离传输的全球性有毒重金属[1-4]. 汞具有高挥发性、生物累积性和持久性的特点,对人类健康和环境造成严重危害[5-6]. 汞可通过自然过程(如岩石风化、地热活动和火山爆发)和人类活动(如采矿、工业生产和化石燃料燃烧)排放进入大气[7-9]. 海洋在全球汞循环中发挥着重要作用[10]. 2018年联合国环境规划署报告指出,每年排放到大气中的汞约有3700 t会沉降到海洋[11]. 自工业革命以来,随着人为排放汞的增加,海洋中汞浓度显著增加[12],导致海洋鱼类体内汞浓度升高,对人类健康造成巨大威胁[13-14]. 海洋环境中不同形态的汞在迁移转化过程中始终处于一个动态平衡状态,主要控制过程包括甲基化/去甲基化、吸附/解吸、沉积物-水界面交换、氧化/还原等(图1). 其中,吸附/解吸、沉积物-水界面交换等迁移过程控制着汞在不同介质中的分配,氧化/还原过程控制着水体中Hg0的水平,而甲基化/去甲基化是控制海洋环境中甲基汞循环的关键过程.
汞在环境中的主要赋存形态包括:Hg0、Hg2+和MeHg[15-19]. MeHg是一种毒性强、易富集、具有严重生态风险的有毒污染物[5, 20-22],这种生物可利用性高的汞形态能够在高营养级生物体内累积到较高浓度水平[20-21, 23]. 食用海产品是人类暴露MeHg的主要途径,可以导致神经发育障碍和自身免疫反应等疾病[20-21, 24-25]. 通过各种来源排放到海洋环境中的无机汞在沉积物和水柱中均可甲基化为MeHg. 近海沉积物中产生的MeHg也可通过长距离传输进入大洋. 原位生成是海洋甲基汞的主要来源,原位甲基化和去甲基化被认为是控制海洋环境中甲基汞水平的重要过程[18-19, 26-27]. 汞的甲基化和去甲基化均可以通过生物和化学(光化学和非光化学)途径发生[28-29],微生物途径被认为是海洋生态系统中甲基汞生成降解的主要途径[30-33]. 光化学途径在表层海水汞迁移转化过程中发挥着重要作用[34-35].
目前已有综述对环境汞的甲基化与去甲基化过程进行了总结[36-39],但对海洋环境中汞甲基化/去甲基化涉及相对较少. 本文在总结海洋汞甲基化/去甲基化途径、过程机制和微生物类群等基础上,详细讨论了海洋微生物(细菌、古菌和微藻)、海洋光化学以及海洋非光化学汞甲基化/去甲基化过程机制.
1. 海洋汞甲基化/去甲基化速率与热点区域(Rates and hotspots of mercury methylation/demethylation in the marine environment)
1.1 海洋汞甲基化/去甲基化过程速率
稳定同位素添加技术具有较高的准确度、精密度和可同时测定甲基化(km)和去甲基化速率(kd)的优点,是测定汞甲基化/去甲基化速率的有效工具[40-41]. 近年来,该技术已被广泛应用于MeHg净产量的估算,以解决水生系统中MeHg的原位生成和控制因素等问题[42-43]. 表1总结了目前报道的稳定同位素添加技术测得的海洋体系中沉积物和水体汞甲基化与去甲基化速率,其中沉积物km范围为0.001—0.21 d−1,kd为0.02—6.4 d−1;水体km范围为0.001—0.005 d−1,kd为0.09—0.57 d−1. 大洋与近岸水体比较,近岸水体汞甲基化与去甲基化速率较高;海洋体系与淡水体系比较发现,海洋体系沉积物中汞甲基化和去甲基化速率更高.
表 1 海洋和淡水系统汞甲基化和去甲基化速率Table 1. Mercury methylation and demethylation rates in various marine and freshwater systems研究区域Study area 介质类型Matrices 甲基化速率/d−1km 去甲基化速率/d−1kd 参考文献Reference 海洋 Marine 长岛海峡 沉积物 0.014—0.082 a 2.0—20 a [44] 新英格兰海岸大陆架 沉积物 0.02—0.21 a — [45] 大西洋中部大陆架 沉积物 0.002—0.053 a 0.02—0.04 a [46] 大西洋中部斜坡 沉积物 0.02—0.05 a 0.4—6.4 a [46] 墨西哥湾 沉积物 0.02—0.19 a 1.6—2.6 a [47] 切萨皮克湾 沉积物 0.01—0.1 a <1—16 a [48-49] 芬迪湾 沉积物 0.0011—0.0018 a 0.15—0.24 a [50] 旧金山湾 沉积物 0.0003—0.006 a — [51] 赤道太平洋 水体 0.004—0.02 a ND—0.78 a [52] 0.002—0.06 b ND—0.55 b 地中海 水体 0.003—0.04 a 0.064—0.19 a [53] <0.0002 b 0.033—0.064 b <0.0002—0.03 c 0.028—0.084 c 淡水 Freshwater 阿杜尔河 沉积物水体 0.03 a 0.096 a [54] <0.0001—0.005 a 0.031—0.903 a [55] <0.0001—0.004 b <0.02—0.576 b <0.0001—0.002 c 0.059—0.0311 c 哈德逊河 沉积物 0.0001—0.0004 a 0.09—1.59 a [50] 大盐湖 沉积物水体 0.0000012—0.0031 a0.0000012—0.0011a — [56] 泻湖(法国) 水体 0.008—0.063 a 0.128—0.245 a [53] 0.002—0.017 b 0.01—0.14 b 0.012—0.031 c 0.01—0.11 c 帕塔克森特河 沉积物 0.0004 a — [50] a代表原位汞甲基化/去甲基化速率;b代表非生物汞甲基化/去甲基化速率;c代表微生物汞甲基化/去甲基化速率. a represents the in situ mercury methylation/demethylation rate; b represents the abiotic mercury methylation/demethylation rate; c represents the microbial mercury methylation/demethylation rate. 1.2 海洋汞甲基化/去甲基化热点区域
沉积物被认为是产生MeHg最重要的场所[57]. 一项在北大西洋的研究表明,硫循环驱动了从近海到深海峡谷沉积物汞的甲基化过程,硫酸盐还原菌(sulfate-reducing bacteria,SRB)是主要的汞甲基化微生物[58]. 南海沉积物汞甲基化过程实验结果证实,微生物甲基化是该研究区域汞甲基化的重要途径,硫酸盐还原菌和铁还原菌(iron- reducing bacteria,FeRB)是主要的甲基化微生物[59]. 汞去甲基化也广泛地发生在墨西哥湾北部、长岛海峡、大西洋中部大陆架和斜坡等海洋沉积物环境中[44-47]. 由此可见,海洋沉积物环境是汞甲基化/去甲基化的重要热点区域.
除沉积物外,汞甲基化/去甲基化过程也可能发生在海洋系统的水柱中. 最近研究表明,海洋次表层可能是汞甲基化的一个潜在热点区域. 例如,研究发现北冰洋大部分MeHg净生产发生在次表层[60]. 萨尼奇湾汞甲基化功能基因hgcAB在200 m深度的样品中丰度最高,表明潜在的新型海洋汞甲基化微生物可能具有更强的耐氧能力和更广的栖息范围[30]. 开阔大洋汞甲基化过程的研究提出,含氧水柱中的好氧微生物可能是次表层汞甲基化的主要微生物[61-64]. 与之相反,在赤道北太平洋次表层溶解氧最小区的研究发现,厌氧产甲烷菌是导致水柱中甲基汞浓度升高的主要微生物[65]. 海洋生物汞同位素组成测定结果也表明汞甲基化过程可能主要发生在浅层海水,即50—400 m深度之间[31, 66]. 太平洋中部汞甲基化和去甲基化的研究揭示,非生物甲基化/去甲基化过程在控制寡营养区域次表层水体甲基汞浓度方面发挥着重要作用[52]. 海底热液系统可能释放Hg和MeHg,因此也被认为是海洋中MeHg的潜在重要来源. 例如,戈尔达海岭(Gorda Ridge)的热液喷口流体中存在高比例的MeHg,研究者认为这可能是由系统内汞的非生物甲基化产生[67]. 微生物宏基因组结果也表明海底热液存在具有汞甲基化潜能的嗜热古菌Pyrococcus furiosus,可能是潜在的甲基化热点区域[64]. 热液喷口和地热温泉等高温环境中微生物去甲基化过程也较为活跃. 微生物去甲基化最早的分支来自嗜热细菌Aquificales,与该菌的栖息地——热液环境中高浓度的Hg2+一致[68]. 随着宏基因组学和单细胞基因组学等测序技术在海洋环境中的广泛应用,已有6个门的去甲基化微生物在热液和地热泉等环境中被鉴定出来[69].
2. 海洋汞光化学甲基化/去甲基化过程机制(Mechanisms of mercury photochemical methylation/demethylation in the marine environment)
人们普遍认为汞甲基化主要由微生物介导,但如果存在合适的甲基供体,无机汞也可以通过非生物途径发生甲基化反应[33, 70]. 根据是否需要光照,非生物汞甲基化分为光化学和非光化学甲基化[18]. 非生物汞甲基化过程作为汞在氧化海水中甲基化的可能途径,对海洋水体MeHg的生成也有一定贡献[27, 52-53]. 太平洋中部的一项研究发现,相对于未过滤海水,过滤海水中汞甲基化速率更高[52]. 由于海水中大部分微生物已通过0.2 μm滤膜去除掉,因此过滤海水中汞甲基化能力增强最有可能归因于非生物机制,而不是由微生物汞甲基化所引起[52]. 此外,同位素示踪实验研究发现,加入同位素标记的无机汞后会立即发生汞的甲基化,这种现象只能通过非生物汞甲基化来解释[27, 52-53, 71-72]. 非生物汞甲基化在海洋环境中的作用也得到最近一项模拟全球海洋汞甲基化过程研究的支持。该研究表明,如果不考虑非生物汞甲基化,模拟的北冰洋和南大洋甲基汞浓度将被大大低估[73].
2.1 海洋汞光化学甲基化过程机制
研究表明,可能促进海洋汞非生物甲基化的化合物包括碘甲烷(CH3I)和二甲基硫(DMS)等. CH3I在海洋环境中由蓝藻等藻类或真菌产生,而DMS主要由海洋浮游植物产生[70, 74]. 有研究表明CH3I诱导的光化学汞甲基化可以在水生环境发生,但该研究设置的CH3I的浓度(mmol·L−1水平)远高于海水(pmol·L−1水平)中的实际浓度,该过程在海水中的贡献尚有待验证[70, 75]. 此外,也有研究表明溶解有机质(DOM)驱动的光化学汞甲基化在降水和湖泊MeHg生成中起着重要作用,但其在海洋环境中的作用尚不清楚[76-77]. 目前普遍认为光化学汞甲基化不太可能成为海洋中甲基汞的主要来源,海洋表层水体MeHg浓度总体较低也支持了这一观点[19].
2.2 海洋汞光化学去甲基化过程机制
生物去甲基化在海水(尤其是深层水体)和沉积物MeHg降解中起着重要作用. 非生物去甲基化也可发生,且大部分研究集中在光化学去甲基化上,这一过程对海洋MeHg降解的贡献可以达到56%—80%[78-79]. MeHg可以通过直接或间接的光化学反应进行降解. 直接光降解利用海水中MeHg的C—Hg键或MeHg-DOM络合物的Hg—S键吸收光,导致键的直接能量转移和断裂[14, 80]. 间接光降解一般归因于光化学诱导产生的活性氧和自由基(如过氧化氢、羟基自由基、单线态氧、过氧化物和DOM的激发三重态)对MeHg的降解[79, 81-82]. 光强、光波长以及水体的理化参数等环境因素均对海水甲基汞光降解产生影响.
赤道太平洋、北冰洋和白令海峡等海域的MeHg光化学去甲基化现场培养实验表明,MeHg降解速率与光强度和波长之间存在明显的相关性[26]. 一般认为,紫外线(UV)是导致MeHg光降解的主要光波段,波长范围为280 nm至400 nm的UV-A和UV-B可以显著影响MeHg的光降解过程[79, 81, 83-84]. 利用汞同位素的非质量分馏(Mass-independent fractionation,MIF)特征示踪海洋生物MeHg的来源时发现,MIF与太阳辐射能量之间密切相关,光化学去甲基化是太平洋表层水中甲基汞降解的重要途径[66, 85]. 除光谱影响外,海水中的DOM也可以通过多种方式影响MeHg的光降解速率。例如,通过形成MeHg-DOM络合物或光化学反应产生自由基促进MeHg去甲基化,或者通过淬灭自由基或吸收太阳辐射降低光化学去甲基化速率[14, 79-80, 86].
3. 海洋微生物汞甲基化/去甲基化过程机制(Mechanisms of microbial mercury methylation/demethylation in the marine environment)
如图1所述,海洋环境中MeHg的水平受到甲基化和去甲基化过程的共同控制[42, 87],其中微生物过程是甲基汞生成降解的最主要途径[33, 87-88].
3.1 微生物汞甲基化过程机制
微生物汞甲基化可以将海洋环境中的无机汞转化为甲基汞,了解微生物汞甲基化的生化机制至关重要[63-64]. 在发现hgcAB基因对汞甲基化的重要作用后,乙酰辅酶A途径被认为是微生物汞甲基化的主要生化机制[89]. 基于SRB和FeRB两种模型菌株,研究者提出hgcA基因能够编码一种起催化甲基转移作用的类咕啉蛋白(HgcA),而hgcB基因会编码一个与HgcA相关的铁氧化还原蛋白(HgcB),两种蛋白能将Hg通过酶催化的甲基化反应生成MeHg[89-90]. hgcAB基因是汞甲基化能力的预测因子,hgcAB基因的发现有助于人们在不同海洋环境中寻找潜在的汞甲基化微生物[57, 64]. 迄今为止,经实验培养验证的海洋环境中汞甲基化微生物主要来自δ-变形菌门(SRB、FeRB和互营养菌)、厚壁菌门和古菌门(产甲烷菌)的厌氧菌[91]. 海洋环境中已确定为汞甲基化微生物或具有 hgcAB同源物的微生物如表 2 所示。
表 2 海洋环境中已确定为汞甲基化微生物或具有hgcAB同源物的微生物Table 2. Microorganisms identified as either methylators or having the hgcAB homologue in the marine environment门Phylum 纲Class 属Genus 参考文献Reference 广古菌门(古菌)Euryarchaeota 甲烷微菌纲Methanomicrobia 甲烷微菌属Methanoregula [94] 甲基甲烷菌属Methanomethylovorans [57] 甲烷叶菌属Methanolobus [57] 甲烷螺菌属 Methanospirillum [59] 变形菌门(细菌)Proteobacteria δ-变形菌纲Deltaproteobacteria 地杆菌属Geobacter [95] 脱硫碱螺旋菌属Desulfonatronospira [57] 脱硫微菌属Desulfomicrobium [96] 互营菌属Syntrophus [95] 脱硫弧菌属Desulfovibrio [97] 脱硫叶菌属Desulfobulbus [59] 脱硫球菌属Desulfococcus [59] 脱硫单胞菌属Desulfuromonas [95] 脱硫杆菌属Desulfobacterium [59] 脱硫葡萄状菌属Desulfacinum [98] 硝化刺菌属Nitrospina [61-62] 厚壁菌门(细菌)Firmicutes 梭菌纲Clostridia 脱硫弯曲孢菌属Desulfosporosinus [59] 脱亚硫酸杆菌属Desulfitobacterium [59] 还原硫素杆菌属Dethiobacter [59] 醋线菌属Acetonema [59] 产乙醇菌属Ethanoligenens [59] 目前在南大洋、大西洋、西太平洋、赤道太平洋、东海和里海等许多海洋环境的沉积物和水体中均检测到hgcAB基因和汞甲基化微生物的存在[64, 92]. 萨尼奇湾的研究发现,携带hgcAB基因的δ-变形菌在200 m深度的海水中具有较高的丰度[30]. 在南海获得的沉积物样品中检测到的汞甲基化细菌属于3个微生物门,包括δ-变形菌门、厚壁菌门和广古菌门,其中δ-变形菌所携带的hgcAB基因丰度最高,是最主要的汞甲基化微生物[59]. 北冰洋宏基因组中检测到具有hgcAB基因的SRB脱硫杆菌科的序列,赤道北太平洋宏基因组中则鉴定出来自产甲烷菌所携带的hgcAB基因[65]. 在波罗的海的缺氧水体和海洋雪中也发现hgcAB基因,系统发育分析表明缺氧水中δ-变形菌是最主要的汞甲基化微生物[93]. 虽然目前已鉴定的汞甲基化菌株多为厌氧菌,但好氧微生物对海洋汞甲基化过程的贡献也不能忽视[64]. 例如,携带hgcAB基因的海洋微嗜氧硝化刺菌((Nitrospina)被认为是南极海冰、西北太平洋和北冰洋等海洋环境中潜在的汞甲基化微生物[61-63, 65].
微生物汞甲基化是一个复杂的过程,可能会受到温度、pH、氧化还原电位(Eh)、DOM等各种生物地球化学因素的影响[99]. 温度升高通常会刺激微生物活动,导致汞甲基化速率增加. 在北极沿海沉积物的研究中发现温度升高会导致SRB活性增强,甲基化速率也随之增加[100]. 研究报道低pH值可以促进甲基汞的生成:一方面是因为Hg2+生物可利用度随着pH值的降低而增加;另一方面是因为汞甲基化微生物在较低pH值下能占据主导地位,从而促进甲基化过程的发生[101]. 一项在波罗的海的研究表明,海洋沉积物中微生物甲基化速率与Eh存在负相关关系[102]. 在影响汞甲基化速率的所有生物地球化学因素中,DOM被认为是最重要的因素之一[103]. DOM可以通过两种方式影响汞的生物可利用性和甲基化潜力[99]:一方面,Hg与DOM形成的络合物难以通过细胞膜扩散,从而降低汞的生物可利用性[104];另一方面,许多研究发现沉积物中MeHg浓度与DOM含量具有较好的正相关关系,这可能是由于DOM作为生长所需的营养物质促进了汞甲基化微生物的生长[105].
3.2 微生物汞去甲基化过程机制
微生物去甲基化是MeHg在海洋水体和沉积物中进行降解的主要途径[23, 106-107]. 绝大多数被鉴定为能发生汞甲基化过程的微生物也具有降解MeHg的能力[96, 108]. 因此,在总体评估海洋环境中汞甲基化潜力时,应同时考虑微生物对MeHg的去甲基化作用.
3.2.1 还原去甲基化过程机制
研究表明微生物降解甲基汞具有两种途径:第一种为mer操纵子系统介导的还原去甲基化,导致Hg0和CH4的生成[107, 109-114]; 第二种为氧化去甲基化,MeHg被降解为Hg2+、CO2和少量CH4[18, 23, 108]. mer体系广泛分布于原核生物中,利用Hg0的低水溶性和易挥发的特点,原核生物可以将Hg2+还原为Hg0,使其能够在高汞浓度下生长[106, 115]. 该系统由汞还原酶(MerA)和多种运输功能蛋白质组成,并受到MerR的精细调控[106, 115]. MerR既是功能性基因表达的抑制因子,也是功能性基因表达的激活因子[106-107, 109-111, 114]. 编码这些不同功能的基因集中在主操纵子中[106-107, 110]. 少数抗汞细菌,即所谓的广谱抗汞细菌,除了对Hg2+有抗性外,还对有机汞化合物有抗性,并编码有机汞裂解酶(MerB)[112-113, 116-119].
参与MeHg还原去甲基化的微生物所拥有的mer操纵子主要位于质粒、染色体、转座子和整合素中,这意味着mer操纵子可以很容易地转移到其他微生物中,因此mer操纵子分布广泛[107]. 在南极海冰、红海的深海和热盐水环境中均检测到merA基因的存在[69],merB基因也在赤道附近的北太平洋和北冰洋水域被检测到[30, 65, 120-121]. 研究发现赤道北太平洋溶解氧最小区mer基因的相对丰度超过了hgcA基因的相对丰度,与该区域水层观测到的MeHg净去甲基化现象一致[65]. 在萨尼奇海湾,merB基因的相对丰度在200 m深处达到峰值,约占微生物群落总基因的0.6%,同时也观察到随merB基因丰度的增加MeHg水平下降的现象[30]. 目前,已在不同海域分离鉴定出290种具有还原去甲基化能力的海洋异养细菌,其中244种被归类为Alteromonas属,46种是Marinobacter属[122].
3.2.2 氧化去甲基化过程机制
与mer操纵子介导的还原去甲基化过程相比,氧化去甲基化过程的研究相对较少[106]. 氧化去甲基化是一个非特异性的代谢过程,与汞的解毒无关,在缺氧环境中常见[106-107, 123-126]. 虽然缺乏纯培养体系对氧化去甲基化过程的验证,但SRB和产甲烷菌仍被认为是其主要参与者[106-108, 114]. 氧化去甲基化过程之所以被如此命名,是因为去甲基化的最终产物是氧化形式的Hg2+、CO2和少量的CH4[23, 108, 124-125]. SRB和产甲烷菌参与氧化去甲基化的区别在于前者可以产生H2S和CO2(方程1),而后者产生CH4和CO2(方程2)[23, 106, 124, 126]. 对于这种现象,研究者推测可能与SRB和产甲烷菌的一碳(C1)代谢有关[23, 106, 108, 124, 126]. 与携带MerB的微生物的活性解毒机制不同,氧化去甲基化是微生物代谢的一部分,甲基汞可能作为电子供体产生作用[87, 106-107, 126]. 据推测,SRB降解MeHg的机制与乙酸盐氧化的机制相似(方程1),而产甲烷菌对MeHg的共同代谢则通过类似于一甲胺降解的途径进行(方程2)[106-107, 115]:
stringUtils.convertMath(!{formula.content}) (1) stringUtils.convertMath(!{formula.content}) (2) 虽然这些假设的氧化去甲基化途径是合理的,但尚未得到实验验证. 海洋和沉积物中氧化降解甲基汞的细菌菌株尚未被分离或鉴定,氧化去甲基化的途径仍有待深入研究[106, 108, 114].
3.3 海洋微藻汞甲基化/去甲基化过程机制
海洋微藻是海洋生态系统中最重要的初级生产者,其对汞的吸收和分子转化是海洋中汞生物积累的重要过程[127-128]. 目前仅有少数研究探究了海洋微藻对汞的甲基化和去甲基化作用,且不同研究的结果存在矛盾[127, 129-130]. 早期研究发现海洋硅藻三角褐指藻能在高浓度HgCl2 (20—120 μg·L−1)条件下诱导Hg的甲基化,作者们认为该过程发生在藻细胞内部,并向外界环境释放MeHg[129]. 但由于实验中添加的汞含量远高于其在天然海水中的浓度,对于海洋微藻是否参与自然海洋环境中汞的甲基化过程仍然缺乏明确的认识. 一项利用同位素示踪技术测定绿藻汞甲基化能力的研究表明,在模拟自然海水条件下莱茵衣藻并不能导致汞的甲基化[131]. 最近我们测试了4门15种海洋微藻的汞甲基化能力,发现被测微藻均未显示出对无机汞的甲基化能力,表明浮游植物在海洋环境甲基汞生成中的作用可能可以忽略不计[132]. 三项研究结果的差异可能是由于高浓度Hg2+条件下,微藻分泌的由脂肪酸和多肽等物质组成的DOM会引起汞的化学甲基化反应[132-134].
相较于机制不明的甲基化过程,有研究发现藻细胞介导的可见光对MeHg的去甲基化可能占水体光化学驱动的去甲基化总量的20%—55%[130],在甲基汞降解中起着重要作用. 放射性同位素(CH3203Hg)实验结果表明,MeHg可以被球石藻降解为Hg0[127]. 同位素示踪研究发现莱茵衣藻能导致MeHg降解,藻细胞内合成的谷胱甘肽以及植物螯合肽可以极大地促进MeHg的去甲基化过程[129]. 最近我们测试了15种常见海洋微藻对汞的去甲基化能力,发现微藻胞外分泌物引起的MeHg光去甲基化速率在0.01—0.39 d−1之间,且部分微藻的共生菌也能诱导MeHg的去甲基化(0.03—0.14 d−1)[132]. 微藻胞外分泌物介导的MeHg光去甲基化速率与现场MeHg光去甲基化和微生物去甲基化速率相当,表明微藻在海洋环境MeHg的去甲基化中发挥着重要作用. 总结可知,藻类介导的MeHg的去甲基化可能主要是由其胞外分泌物(通过光介导途径)引起的,而不是微藻细胞对MeHg的直接去甲基化[127, 131-132]. 进一步研究也发现巯基可能是微藻胞外分泌物中导致MeHg光去甲基化的主要成分[132, 135].
4. 海洋非光化学汞甲基化/去甲基化过程机制(Mechanism of non-photochemical mercury methylation/demethylation in the marine environment)
4.1 海洋非光化学汞甲基化过程机制
非光化学Hg甲基化包括以有机金属配合物作为甲基供体的转甲基化[136],以甲基钴胺素为甲基供体的细胞外汞甲基化[137],以及腐殖质对汞的甲基化[138]. 目前已经证明在海洋环境中除了甲基锡之外,其余各种有机金属化合物的转甲基化途径都无关紧要[70, 139]. 然而,Celo等研究使用的甲基锡浓度远高于海水中甲基锡的测量值,因此甲基锡化合物介导的汞非生物甲基化可能也并不是海水中甲基汞的主要来源[70]. 细胞外汞甲基化主要指由生物体释放到环境中的甲基钴胺素导致的非酶化汞甲基化,由此将其与hgcAB介导的细胞内汞甲基化(也涉及甲基钴胺素)区分开来[89]. 研究发现在甲基钴胺素介导的细胞外汞甲基化过程中,Hg2+作为亲电试剂攻击甲基钴胺素,导致解离出CH3−,随后CH3−转移到高氧化态的Hg2+上生成MeHg[140-141]. 对于腐殖质介导的汞化学甲基化,CH3−向Hg2+的转移被认为是主要的甲基化途径[134]. 由于腐殖质的组成不同,汞甲基化所涉及的化合物组分不同,反应动力学也存在差异,并且对腐殖质中特定甲基供体的汞甲基化缺乏了解,因此尚无法定量估计其对海水中MeHg的贡献[138, 142]. 总体而言,虽然很难估计非生物汞甲基化对海水MeHg的贡献,但已有的研究表明,甲基钴胺素和腐殖质介导的汞甲基化在海水中可能起着一定作用[52, 73].
4.2 海洋非光化学汞去甲基化过程机制
虽然大多数关于MeHg降解的研究主要集中在光化学和微生物去甲基化,但非光化学去甲基化也已被证实可以在水环境中发生. 目前有两种化学去甲基化途径已在研究中得到证实:(1)是甲基汞与H2S或硫化物矿物反应生成HgS(s)和二甲基汞(DMeHg)[143-145];(2)是MeHg与硒氨基酸反应生成HgSe(s)[146-147].
最近的研究证明,MeHg吸附在硫化物矿物或有机二硫醇表面会形成DMeHg[143-144]. 硫化物矿物上多个S原子与Hg2+进行配位形成稳定的过渡态,并激活C—Hg键以进行甲基转移或形成DMeHg[143, 148-149]. MeHg和H2S之间的反应通过强的Hg—Hg相互作用激活甲基在分子内转移,诱导(CH3Hg)2S复合体的重排或甲基直接从一个取代基转移到另一个取代基,从而促进形成DMeHg[148]. 这些重要的发现表明矿物表面的硫基减少可以导致MeHg的降解,并伴随海洋环境中DMeHg的生成[143-144]. 与硫化物介导的去甲基化途径类似,硒介导的去甲基化被认为涉及MeHg和硒氨基酸之间的反应,通过形成(CH3Hg)2Se和DMeHg作为中间体,最终降解产物为HgSe(s)[146-147]. 虽然这些反应是在相对较高的硒水平下观察到的,但这种DMeHg去甲基化的机制已经在海洋哺乳动物的肝脏和肠道中被检测到[150-151]. 这些结果表明,硒可以将MeHg转化为无机汞,从而降低其生物可利用性和生物积累.
5. 展望(Prospect)
(1)不同汞甲基化/去甲基化途径贡献估算:汞甲基化/去甲基化包括微生物、光化学和非光化学等多种途径,量化各途径贡献是认识MeHg迁移转化循环的基础,改进汞同位素分馏技术并结合同位素添加示踪技术有望实现区分非生物与生物甲基化/去甲基化相对贡献的目标.
(2)实际海洋环境汞甲基化/去甲基化基因/微生物作用验证:虽然目前已有一些关于海洋环境中汞甲基化/去甲基化微生物和功能基因的初步报道,但对这一过程中关键微生物和功能基因分布及其在海洋汞循环过程中的作用仍缺乏基本了解, 解决这一问题将有助于理解hgcAB和merAB等功能基因及相关微生物在实际海洋环境中的作用,并为预测海洋系统中汞的长期风险提供数据支撑.
(3)定量环境因素对海洋汞甲基化/去甲基化的影响:海洋汞甲基化/去甲基化过程受到多种物理、化学与生物因素的影响,包括温度、Eh、pH、无机汞的形态和浓度、DOM和微生物群落结构等. 定量这些环境因素对海洋汞甲基化/去甲基化的影响是预测未来气候变化和生态系统演变背景下汞长期风险的关键.
-
表 1 海洋和淡水系统汞甲基化和去甲基化速率
Table 1. Mercury methylation and demethylation rates in various marine and freshwater systems
研究区域Study area 介质类型Matrices 甲基化速率/d−1km 去甲基化速率/d−1kd 参考文献Reference 海洋 Marine 长岛海峡 沉积物 0.014—0.082 a 2.0—20 a [44] 新英格兰海岸大陆架 沉积物 0.02—0.21 a — [45] 大西洋中部大陆架 沉积物 0.002—0.053 a 0.02—0.04 a [46] 大西洋中部斜坡 沉积物 0.02—0.05 a 0.4—6.4 a [46] 墨西哥湾 沉积物 0.02—0.19 a 1.6—2.6 a [47] 切萨皮克湾 沉积物 0.01—0.1 a <1—16 a [48-49] 芬迪湾 沉积物 0.0011—0.0018 a 0.15—0.24 a [50] 旧金山湾 沉积物 0.0003—0.006 a — [51] 赤道太平洋 水体 0.004—0.02 a ND—0.78 a [52] 0.002—0.06 b ND—0.55 b 地中海 水体 0.003—0.04 a 0.064—0.19 a [53] <0.0002 b 0.033—0.064 b <0.0002—0.03 c 0.028—0.084 c 淡水 Freshwater 阿杜尔河 沉积物水体 0.03 a 0.096 a [54] <0.0001—0.005 a 0.031—0.903 a [55] <0.0001—0.004 b <0.02—0.576 b <0.0001—0.002 c 0.059—0.0311 c 哈德逊河 沉积物 0.0001—0.0004 a 0.09—1.59 a [50] 大盐湖 沉积物水体 0.0000012—0.0031 a0.0000012—0.0011a — [56] 泻湖(法国) 水体 0.008—0.063 a 0.128—0.245 a [53] 0.002—0.017 b 0.01—0.14 b 0.012—0.031 c 0.01—0.11 c 帕塔克森特河 沉积物 0.0004 a — [50] a代表原位汞甲基化/去甲基化速率;b代表非生物汞甲基化/去甲基化速率;c代表微生物汞甲基化/去甲基化速率. a represents the in situ mercury methylation/demethylation rate; b represents the abiotic mercury methylation/demethylation rate; c represents the microbial mercury methylation/demethylation rate. 表 2 海洋环境中已确定为汞甲基化微生物或具有hgcAB同源物的微生物
Table 2. Microorganisms identified as either methylators or having the hgcAB homologue in the marine environment
门Phylum 纲Class 属Genus 参考文献Reference 广古菌门(古菌)Euryarchaeota 甲烷微菌纲Methanomicrobia 甲烷微菌属Methanoregula [94] 甲基甲烷菌属Methanomethylovorans [57] 甲烷叶菌属Methanolobus [57] 甲烷螺菌属 Methanospirillum [59] 变形菌门(细菌)Proteobacteria δ-变形菌纲Deltaproteobacteria 地杆菌属Geobacter [95] 脱硫碱螺旋菌属Desulfonatronospira [57] 脱硫微菌属Desulfomicrobium [96] 互营菌属Syntrophus [95] 脱硫弧菌属Desulfovibrio [97] 脱硫叶菌属Desulfobulbus [59] 脱硫球菌属Desulfococcus [59] 脱硫单胞菌属Desulfuromonas [95] 脱硫杆菌属Desulfobacterium [59] 脱硫葡萄状菌属Desulfacinum [98] 硝化刺菌属Nitrospina [61-62] 厚壁菌门(细菌)Firmicutes 梭菌纲Clostridia 脱硫弯曲孢菌属Desulfosporosinus [59] 脱亚硫酸杆菌属Desulfitobacterium [59] 还原硫素杆菌属Dethiobacter [59] 醋线菌属Acetonema [59] 产乙醇菌属Ethanoligenens [59] -
[1] AKSENTOV K I, KALINCHUK V V. Atomic mercury distribution features in the surface air layer in the sea of Japan in the fall of 2010 [J]. Russian Meteorology and Hydrology, 2012, 37(10): 674-680. doi: 10.3103/S1068373912100056 [2] CI Z J, ZHANG X S, WANG Z W, et al. Distribution and air-sea exchange of mercury (Hg) in the Yellow Sea [J]. Atmospheric Chemistry and Physics, 2011, 11(6): 2881-2892. doi: 10.5194/acp-11-2881-2011 [3] SHEU G R, LIN N H, WANG J L, et al. Temporal distribution and potential sources of atmospheric mercury measured at a high-elevation background station in Taiwan [J]. Atmospheric Environment, 2010, 44(20): 2393-2400. doi: 10.1016/j.atmosenv.2010.04.009 [4] JAFFE D, PRESTBO E, SWARTZENDRUBER P, et al. Export of atmospheric mercury from Asia [J]. Atmospheric Environment, 2005, 39(17): 3029-3038. doi: 10.1016/j.atmosenv.2005.01.030 [5] RICE K M, WALKER E M Jr, WU M Z, et al. Environmental mercury and its toxic effects [J]. Journal of Preventive Medicine and Public Health, 2014, 47(2): 74-83. doi: 10.3961/jpmph.2014.47.2.74 [6] DRISCOLL C T, MASON R P, CHAN H M, et al. Mercury as a global pollutant: Sources, pathways, and effects [J]. Environmental Science & Technology, 2013, 47(10): 4967-4983. [7] CAI X R, CAI B F, ZHANG H R, et al. Establishment of high-resolution atmospheric mercury emission inventories for Chinese cement plants based on the mass balance method [J]. Environmental Science & Technology, 2020, 54(21): 13399-13408. [8] XU J Y, BRAVO A G, LAGERKVIST A, et al. Sources and remediation techniques for mercury contaminated soil [J]. Environment International, 2015, 74: 42-53. doi: 10.1016/j.envint.2014.09.007 [9] HOROWITZ H M, JACOB D J, AMOS H M, et al. Historical Mercury releases from commercial products: Global environmental implications [J]. Environmental Science & Technology, 2014, 48(17): 10242-10250. [10] BATRAKOVA N, TRAVNIKOV O, ROZOVSKAYA O. Chemical and physical transformations of mercury in the ocean: A review [J]. Ocean Science, 2014, 10(6): 1047-1063. doi: 10.5194/os-10-1047-2014 [11] UNEP. Global Mercury Assessment 2018: Source, emissions, release, and environmental transport[R]. 2018. [12] LAMBORG C H, HAMMERSCHMIDT C R, BOWMAN K L, et al. A global ocean inventory of anthropogenic mercury based on water column measurements [J]. Nature, 2014, 512(7512): 65-68. doi: 10.1038/nature13563 [13] LIU M D, CHEN L, WANG X J, et al. Mercury export from mainland China to adjacent seas and its influence on the marine mercury balance [J]. Environmental Science & Technology, 2016, 50(12): 6224-6232. [14] QIAN Y, YIN X P, LIN H, et al. Why dissolved organic matter enhances photodegradation of methylmercury [J]. Environmental Science and Technology Letters, 2014, 1(10): 426-431. doi: 10.1021/ez500254z [15] BECKERS F, RINKLEBE J. Cycling of mercury in the environment: Sources, fate, and human health implications: A review [J]. Critical Reviews in Environmental Science and Technology, 2017, 47(9): 693-794. doi: 10.1080/10643389.2017.1326277 [16] BRAUNE B, CHÉTELAT J, AMYOT M, et al. Mercury in the marine environment of the Canadian Arctic: Review of recent findings [J]. Science of the Total Environment, 2015, 509/510: 67-90. doi: 10.1016/j.scitotenv.2014.05.133 [17] BOWMAN K L, HAMMERSCHMIDT C R, LAMBORG C H, et al. Mercury in the north Atlantic Ocean: The US Geotraces zonal and meridional sections [J]. Deep Sea Research Part II:Topical Studies in Oceanography, 2015, 116: 251-261. doi: 10.1016/j.dsr2.2014.07.004 [18] LI Y B, CAI Y. Progress in the study of mercury methylation and demethylation in aquatic environments [J]. Chinese Science Bulletin, 2013, 58(2): 177-185. doi: 10.1007/s11434-012-5416-4 [19] MASON R P, CHOI A L, FITZGERALD W F, et al. Mercury biogeochemical cycling in the ocean and policy implications [J]. Environmental Research, 2012, 119: 101-117. doi: 10.1016/j.envres.2012.03.013 [20] PETROVA M V, OURGAUD M, BOAVIDA J R H, et al. Human mercury exposure levels and fish consumption at the French Riviera [J]. Chemosphere, 2020, 258: 127232. doi: 10.1016/j.chemosphere.2020.127232 [21] SCHAEFER A M, ZOFFER M, YRASTORZA L, et al. Mercury exposure, fish consumption, and perceived risk among pregnant women in coastal Florida [J]. International Journal of Environmental Research and Public Health, 2019, 16(24): 4903. doi: 10.3390/ijerph16244903 [22] MERGLER D, ANDERSON H A, CHAN L H M, et al. Methylmercury exposure and health effects in humans: A worldwide concern [J]. Ambio, 2007, 36(1): 3-11. doi: 10.1579/0044-7447(2007)36[3:MEAHEI]2.0.CO;2 [23] HSU-KIM H, KUCHARZYK K H, ZHANG T, et al. Mechanisms regulating mercury bioavailability for methylating microorganisms in the aquatic environment: A critical review [J]. Environmental Science & Technology, 2013, 47(6): 2441-2456. [24] SCHAEFER A M, JENSEN E L, BOSSART G D, et al. Hair mercury concentrations and fish consumption patterns in Florida residents [J]. International Journal of Environmental Research and Public Health, 2014, 11(7): 6709-6726. doi: 10.3390/ijerph110706709 [25] KARAGAS M R, CHOI A L, OKEN E, et al. Evidence on the human health effects of low-level methylmercury exposure [J]. Environmental Health Perspectives, 2012, 120(6): 799-806. doi: 10.1289/ehp.1104494 [26] DIMENTO B P, MASON R P. Factors controlling the photochemical degradation of methylmercury in coastal and oceanic waters [J]. Marine Chemistry, 2017, 196: 116-125. doi: 10.1016/j.marchem.2017.08.006 [27] LEHNHERR I, ST LOUIS V L, HINTELMANN H, et al. Methylation of inorganic mercury in polar marine waters [J]. Nature Geoscience, 2011, 4(5): 298-302. doi: 10.1038/ngeo1134 [28] CHEN B W, CHEN P, HE B, et al. Identification of mercury methylation product by tert-butyl compounds in aqueous solution under light irradiation [J]. Marine Pollution Bulletin, 2015, 98(1/2): 40-46. [29] HAMELIN S, AMYOT M, BARKAY T, et al. Methanogens: principal methylators of mercury in lake periphyton [J]. Environmental Science & Technology, 2011, 45(18): 7693-7700. [30] LIN H Y, ASCHER D B, MYUNG Y, et al. Mercury methylation by metabolically versatile and cosmopolitan marine bacteria [J]. The ISME Journal, 2021, 15(6): 1810-1825. doi: 10.1038/s41396-020-00889-4 [31] BLUM J D, POPP B N, DRAZEN J C, et al. Methylmercury production below the mixed layer in the North Pacific Ocean [J]. Nature Geoscience, 2013, 6(10): 879-884. doi: 10.1038/ngeo1918 [32] SELIN N E. Global biogeochemical cycling of mercury: A review [J]. Annual Review of Environment and Resources, 2009, 34: 43-63. doi: 10.1146/annurev.environ.051308.084314 [33] ULLRICH S M, TANTON T W, ABDRASHITOVA S A. Mercury in the aquatic environment: A review of factors affecting methylation [J]. Critical Reviews in Environmental Science and Technology, 2001, 31(3): 241-293. doi: 10.1080/20016491089226 [34] LUO H W, CHENG Q Q, PAN X L. Photochemical behaviors of mercury (Hg) species in aquatic systems: A systematic review on reaction process, mechanism, and influencing factor [J]. Science of the Total Environment, 2020, 720: 137540. doi: 10.1016/j.scitotenv.2020.137540 [35] ROSE C H, GHOSH S, BLUM J D, et al. Effects of ultraviolet radiation on mercury isotope fractionation during photo-reduction for inorganic and organic mercury species [J]. Chemical Geology, 2015, 405: 102-111. doi: 10.1016/j.chemgeo.2015.02.025 [36] 毕丽, 贺纪正, 张丽梅, 等. 环境中汞微生物转化研究进展 [J]. 环境化学, 2018, 37(11): 2359-2367. doi: 10.7524/j.issn.0254-6108.2018011703 BI L, HE J Z, ZHANG L M, et al. Microbial transformations of mercury in the environment [J]. Environmental Chemistry, 2018, 37(11): 2359-2367(in Chinese). doi: 10.7524/j.issn.0254-6108.2018011703
[37] 谷春豪, 许怀凤, 仇广乐. 汞的微生物甲基化与去甲基化机理研究进展 [J]. 环境化学, 2013, 32(6): 926-936. doi: 10.7524/j.issn.0254-6108.2013.06.002 GU C H, XU H F, QIU G L. The progress in research on mechanism of microbial mercury methylation and de-methylation [J]. Environmental Chemistry, 2013, 32(6): 926-936(in Chinese). doi: 10.7524/j.issn.0254-6108.2013.06.002
[38] 胡海燕, 冯新斌, 曾永平, 等. 汞的微生物甲基化研究进展 [J]. 生态学杂志, 2011, 30(5): 874-882. doi: 10.13292/j.1000-4890.2011.0143 HU H Y, FENG X B, ZENG Y P, et al. Progress in research on microbial methylation of mercury [J]. Chinese Journal of Ecology, 2011, 30(5): 874-882(in Chinese). doi: 10.13292/j.1000-4890.2011.0143
[39] 刘金铃, 丁振华. 汞的甲基化研究进展 [J]. 地球与环境, 2007, 35(3): 215-222. doi: 10.3969/j.issn.1672-9250.2007.03.004 LIU J L, DING Z H. Progress in research on mercury methylation in environment [J]. Earth and Environment, 2007, 35(3): 215-222(in Chinese). doi: 10.3969/j.issn.1672-9250.2007.03.004
[40] HINTELMANN H, KEPPEL-JONES K, EVANS R D. Constants of mercury methylation and demethylation rates in sediments and comparison of tracer and ambient mercury availability [J]. Environmental Toxicology and Chemistry, 2000, 19(9): 2204-2211. doi: 10.1002/etc.5620190909 [41] FURUTANI A, RUDD J W. Measurement of mercury methylation in lake water and sediment samples [J]. Applied and Environmental Microbiology, 1980, 40(4): 770-776. doi: 10.1128/aem.40.4.770-776.1980 [42] FIGUEIREDO N, SERRALHEIRO M L, CANARIO J, et al. Evidence of mercury methylation and demethylation by the estuarine microbial communities obtained in stable Hg isotope studies [J]. International Journal of Environmental Research and Public Health, 2018, 15(10): 2141. doi: 10.3390/ijerph15102141 [43] LI Y B, YIN Y G, LIU G L, et al. Estimation of the major source and sink of methylmercury in the Florida Everglades [J]. Environmental Science & Technology, 2012, 46(11): 5885-5893. [44] HAMMERSCHMIDT C R, FITZGERALD W F, LAMBORG C H, et al. Biogeochemistry of methylmercury in sediments of Long Island Sound [J]. Marine Chemistry, 2004, 90(1/2/3/4): 31-52. [45] HAMMERSCHMIDT C R, FITZGERALD W F. Methylmercury cycling in sediments on the continental shelf of southern New England [J]. Geochimica et Cosmochimica Acta, 2006, 70(4): 918-930. doi: 10.1016/j.gca.2005.10.020 [46] HOLLWEG T A, GILMOUR, MASON R P. Mercury and methylmercury cycling in sediments of the mid-Atlantic continental shelf and slope [J]. Limnology and Oceanography, 2010, 55(6): 2703-2722. doi: 10.4319/lo.2010.55.6.2703 [47] LIU B, SCHAIDER L A, MASON R P, et al. Controls on methylmercury accumulation in northern Gulf of Mexico sediments [J]. Estuarine, Coastal and Shelf Science, 2015, 159: 50-59. doi: 10.1016/j.ecss.2015.03.030 [48] HOLLWEG T A. Mercury cycling in sediments of Chesapeake Bay and the mid-Atlantic continental shelf and slope [J]. The Sciences and Engineering Collection, 2010: 3421837. [49] HOLLWEG T A, GILMOUR C C, MASON R P. Methylmercury production in sediments of Chesapeake Bay and the mid-Atlantic continental margin [J]. Marine Chemistry, 2009, 114(3/4): 86-101. [50] HEYES A, MASON R P, KIM E H, et al. Mercury methylation in estuaries: Insights from using measuring rates using stable mercury isotopes [J]. Marine Chemistry, 2006, 102(1/2): 134-147. [51] MARVIN-DIPASQUALE M, AGEE J, BOUSE R, et al. Microbial cycling of mercury in contaminated pelagic and wetland sediments of San Pablo Bay, California [J]. Environmental Geology, 2003, 43(3): 260-267. doi: 10.1007/s00254-002-0623-y [52] MUNSON K M, LAMBORG C H, BOITEAU R M, et al. Dynamic mercury methylation and demethylation in oligotrophic marine water [J]. Biogeosciences, 2018, 15(21): 6451-60. doi: 10.5194/bg-15-6451-2018 [53] MONPERRUS M, TESSIER E, AMOUROUX D, et al. Mercury methylation, demethylation and reduction rates in coastal and marine surface waters of the Mediterranean Sea [J]. Marine Chemistry, 2007, 107(1): 49-63. doi: 10.1016/j.marchem.2007.01.018 [54] RODRı́GUEZ MARTı́N-DOIMEADIOS R C, TESSIER E, AMOUROUX D, et al. Mercury methylation/demethylation and volatilization pathways in estuarine sediment slurries using species-specific enriched stable isotopes [J]. Marine Chemistry, 2004, 90(1/2/3/4): 107-123. [55] SHARIF A, MONPERRUS M, TESSIER E, et al. Fate of mercury species in the coastal plume of the Adour River Estuary (Bay of Biscay, SW France) [J]. Science of the Total Environment, 2014, 496: 701-713. doi: 10.1016/j.scitotenv.2014.06.116 [56] JOHNSON W P, SWANSON N, BLACK B, et al. Total- and methyl-mercury concentrations and methylation rates across the freshwater to hypersaline continuum of the Great Salt Lake, Utah, USA [J]. Science of the Total Environment, 2015, 511: 489-500. doi: 10.1016/j.scitotenv.2014.12.092 [57] GILMOUR C C, PODAR M, BULLOCK A L, et al. Mercury methylation by novel microorganisms from new environments [J]. Environmental Science & Technology, 2013, 47(20): 11810-11820. [58] AZAROFF A, GOÑI URRIZA M, GASSIE C, et al. Marine mercury-methylating microbial communities from coastal to Capbreton Canyon sediments (North Atlantic Ocean) [J]. Environmental Pollution, 2020, 262: 114333. doi: 10.1016/j.envpol.2020.114333 [59] YUAN K, CHEN X, CHEN P, et al. Mercury methylation-related microbes and genes in the sediments of the Pearl River Estuary and the South China Sea [J]. Ecotoxicology and Environmental Safety, 2019, 185: 109722. doi: 10.1016/j.ecoenv.2019.109722 [60] SOERENSEN A L, JACOB D J, SCHARTUP A T, et al. A mass budget for mercury and methylmercury in the Arctic Ocean [J]. Global Biogeochemical Cycles, 2016, 30(4): 560-575. doi: 10.1002/2015GB005280 [61] TADA Y Y, MARUMOTO K, TAKEUCHI A. Nitrospina-like bacteria are dominant potential mercury methylators in both the oyashio and kuroshio regions of the western north Pacific [J]. Microbiology Spectrum, 2021, 9(2): e0083321. doi: 10.1128/Spectrum.00833-21 [62] TADA, MARUMOTO K, TAKEUCHI A. Nitrospina-like bacteria are potential mercury methylators in the mesopelagic zone in the East China Sea [J]. Frontiers in Microbiology, 2020, 11: 1369. doi: 10.3389/fmicb.2020.01369 [63] GIONFRIDDO C M, TATE M T, WICK R R, et al. Microbial mercury methylation in Antarctic Sea ice [J]. Nature Microbiology, 2016, 1: 16127. doi: 10.1038/nmicrobiol.2016.127 [64] PODAR M, GILMOUR C C, BRANDT C C, et al. Global prevalence and distribution of genes and microorganisms involved in mercury methylation [J]. Science Advances, 2015, 1(9): e1500675. doi: 10.1126/sciadv.1500675 [65] BOWMAN K L, COLLINS R E, AGATHER A M, et al. Distribution of mercury-cycling genes in the Arctic and equatorial Pacific Oceans and their relationship to mercury speciation [J]. Limnology and Oceanography, 2019, 65: S310-S320. [66] SUN R Y, YUAN J J, SONKE J E, et al. Methylmercury produced in upper oceans accumulates in deep Mariana Trench fauna [J]. Nature Communications, 2020, 11: 3389. doi: 10.1038/s41467-020-17045-3 [67] LAMBORG C H, von DAMM K L, FITZGERALD W F, et al. Mercury and monomethylmercury in fluids from Sea Cliff submarine hydrothermal field, Gorda Ridge [J]. Geophysical Research Letters, 2006, 33(17): L17606. doi: 10.1029/2006GL026321 [68] BARKAY T, KRITEE K, BOYD E, et al. A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase [J]. Environmental Microbiology, 2010, 12(11): 2904-2917. doi: 10.1111/j.1462-2920.2010.02260.x [69] CHRISTAKIS C A, BARKAY T, BOYD E S. Expanded diversity and phylogeny of mer genes broadens mercury resistance paradigms and reveals an origin for MerA among thermophilic Archaea [J]. Frontiers in Microbiology, 2021, 12: 682605. doi: 10.3389/fmicb.2021.682605 [70] CELO V, LEAN D R S, SCOTT S L. Abiotic methylation of mercury in the aquatic environment [J]. Science of the Total Environment, 2006, 368(1): 126-137. doi: 10.1016/j.scitotenv.2005.09.043 [71] WANG K, MUNSON K M, ARMSTRONG D A, et al. Determining seawater mercury methylation and demethylation rates by the seawater incubation approach: A critique [J]. Marine Chemistry, 2020, 219: 103753. doi: 10.1016/j.marchem.2020.103753 [72] MUNSON K M. Transformations of mercury in the marine water column[D]. Massachusetts: Massachusetts Institute of Technology, 2014. [73] SEMENIUK K, DASTOOR A. Development of a global ocean mercury model with a methylation cycle: Outstanding issues [J]. Global Biogeochemical Cycles, 2017, 31(2): 400-433. [74] SMYTHE-WRIGHT D, BOSWELL S M, BREITHAUPT P, et al. Methyl iodide production in the ocean: Implications for climate change [J]. Global Biogeochemical Cycles, 2006, 20(3): GB3003. [75] YIN Y G, LI Y B, TAI C, et al. Fumigant methyl iodide can methylate inorganic mercury species in natural waters [J]. Nature Communications, 2014, 5: 4633. doi: 10.1038/ncomms5633 [76] HAMMERSCHMIDT C R, LAMBORG C H, FITZGERALD W F. Aqueous phase methylation as a potential source of methylmercury in wet deposition [J]. Atmospheric Environment, 2007, 41(8): 1663-1668. doi: 10.1016/j.atmosenv.2006.10.032 [77] SICILIANO S D, O'DRISCOLL N J, TORDON R, et al. Abiotic production of methylmercury by solar radiation [J]. Environmental Science & Technology, 2005, 39(4): 1071-1077. [78] MOTTA L C, BLUM J D, POPP B N, et al. Mercury stable isotopes in flying fish as a monitor of photochemical degradation of methylmercury in the Atlantic and Pacific Oceans [J]. Marine Chemistry, 2020, 223: 103790. doi: 10.1016/j.marchem.2020.103790 [79] BLACK F J, POULIN B A, FLEGAL A R. Factors controlling the abiotic photo-degradation of monomethylmercury in surface waters [J]. Geochimica et Cosmochimica Acta, 2012, 84: 492-507. doi: 10.1016/j.gca.2012.01.019 [80] JEREMIASON J D, PORTNER J C, AIKEN G R, et al. Photoreduction of Hg(II) and photodemethylation of methylmercury: The key role of thiol sites on dissolved organic matter [J]. Environmental Science. Processes & Impacts, 2015, 17(11): 1892-1903. [81] KIM M K, ZOH K D. Effects of natural water constituents on the photo-decomposition of methylmercury and the role of hydroxyl radical [J]. Science of the Total Environment, 2013, 449: 95-101. doi: 10.1016/j.scitotenv.2013.01.039 [82] ZHANG T, HSU-KIM H. Photolytic degradation of methylmercury enhanced by binding to natural organic ligands [J]. Nature Geoscience, 2010, 3(7): 473-476. doi: 10.1038/ngeo892 [83] GU B, LU X, JOHS A, et al. Encyclopedia of water: science, technology, and society[M]. Wiley, 2019. [84] LI Y B, MAO Y X, LIU G L, et al. Degradation of methylmercury and its effects on mercury distribution and cycling in the Florida Everglades [J]. Environmental Science & Technology, 2010, 44(17): 6661-6666. [85] KIM M K, WON A Y, ZOH K D. The production of dissolved gaseous mercury from methylmercury photodegradation at different salinity [J]. Desalination and Water Treatment, 2016, 57(2): 610-619. doi: 10.1080/19443994.2014.986829 [86] ZHANG D, YIN Y G, LI Y B, et al. Critical role of natural organic matter in photodegradation of methylmercury in water: Molecular weight and interactive effects with other environmental factors [J]. Science of the Total Environment, 2017, 578: 535-541. doi: 10.1016/j.scitotenv.2016.10.222 [87] BRAVO A G, COSIO C. Biotic formation of methylmercury: A bio-physico-chemical conundrum [J]. Limnology and Oceanography, 2020, 65(5): 1010-1027. doi: 10.1002/lno.11366 [88] PARANJAPE A, HALL B. Recent advances in the study of mercury methylation in aquatic systems [J]. Facets, 2017, 2(1): 85-119. doi: 10.1139/facets-2016-0027 [89] PARKS J M, JOHS A, PODAR M, et al. The genetic basis for bacterial mercury methylation [J]. Science, 2013, 339(6125): 1332-1335. doi: 10.1126/science.1230667 [90] POULAIN A J, BARKAY T. Cracking the mercury methylation code [J]. Science, 2013, 339(6125): 1280-1281. doi: 10.1126/science.1235591 [91] GREGOIRE D, POULAIN A. Shining light on recent advances in microbial mercury cycling [J]. Facets, 2018, 3(1): 858-879. doi: 10.1139/facets-2018-0015 [92] VILLAR E, CABROL L, HEIMBÜRGER-BOAVIDA L E. Widespread microbial mercury methylation genes in the global ocean [J]. Environmental Microbiology Reports, 2020, 12(3): 277-287. doi: 10.1111/1758-2229.12829 [93] CAPO E, BRAVO A G, SOERENSEN A L, et al. Marine snow as a habitat for microbial mercury methylators in the Baltic Sea[J]. Biorxiv, 2020,. [94] GILMOUR C C, BULLOCK A L, MCBURNEY A, et al. Robust mercury methylation across diverse methanogenic Archaea [J]. mBio, 2018, 9(2): e02403-e02417. [95] LU X, LIU Y R, JOHS A, et al. Anaerobic mercury methylation and demethylation by Geobacter bemidjiensis bem [J]. Environmental Science & Technology, 2016, 50(8): 4366-4373. [96] BRIDOU R, MONPERRUS M, GONZALEZ P R, et al. Simultaneous determination of mercury methylation and demethylation capacities of various sulfate-reducing bacteria using species-specific isotopic tracers [J]. Environmental Toxicology and Chemistry, 2011, 30(2): 337-344. doi: 10.1002/etc.395 [97] GRAHAM A M, BULLOCK A L, MAIZEL A C, et al. Detailed assessment of the kinetics of Hg-cell association, Hg methylation, and methylmercury degradation in several Desulfovibrio species [J]. Applied and Environmental Microbiology, 2012, 78(20): 7337-7346. doi: 10.1128/AEM.01792-12 [98] MALCOLM E G, SCHAEFER J K, EKSTROM E B, et al. Mercury methylation in oxygen deficient zones of the oceans: No evidence for the predominance of anaerobes [J]. Marine Chemistry, 2010, 122(1/2/3/4): 11-19. [99] MA M, DU H X, WANG D Y. Mercury methylation by anaerobic microorganisms: A review [J]. Critical Reviews in Environmental Science and Technology, 2019, 49(20): 1893-1936. doi: 10.1080/10643389.2019.1594517 [100] ST. PIERRE K A, CH T LAT J, YUMVIHOZE E, et al. Temperature and the sulfur cycle control monomethylmercury cycling in high arctic coastal marine sediments from Allen Bay, Nunavut, Canada [J]. Environmental Science & Technology, 2014, 48(5): 2680-2687. [101] KELLY C A, RUDD J W M, HOLOKA M H. Effect of pH on mercury uptake by an aquatic bacterium: Implications for Hg cycling [J]. Environmental Science & Technology, 2003, 37(13): 2941-2946. [102] BEŁDOWSKI J, MIOTK M, PEMPKOWIAK J. Methylation index as means of quantification of the compliance of sedimentary mercury to be methylated [J]. Environmental Monitoring and Assessment, 2015, 187(8): 498. doi: 10.1007/s10661-015-4716-y [103] ZHAO L D, CHEN H M, LU X, et al. Contrasting effects of dissolved organic matter on mercury methylation by Geobacter sulfurreducens PCA and Desulfovibrio desulfuricans ND132 [J]. Environmental Science & Technology, 2017, 51(18): 10468-10475. [104] GU B, BIAN Y, MILLER C L, et al. Mercury reduction and complexation by natural organic matter in anoxic environments [J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(4): 1479-1483. doi: 10.1073/pnas.1008747108 [105] SCHAEFER J K, ROCKS S S, ZHENG W, et al. Active transport, substrate specificity, and methylation of Hg(II) in anaerobic bacteria [J]. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108(21): 8714-8719. doi: 10.1073/pnas.1105781108 [106] BARKAY T, GU B. Demethylation─The other side of the mercury methylation coin: a critical review [J]. ACS Environmental Au, 2022, 2(2): 77-97. doi: 10.1021/acsenvironau.1c00022 [107] DU H X, MA M, IGARASHI Y, et al. Biotic and abiotic degradation of methylmercury in aquatic ecosystems: A review [J]. Bulletin of Environmental Contamination and Toxicology, 2019, 102(5): 605-611. doi: 10.1007/s00128-018-2530-2 [108] BARKAY T, WAGNER-DÖBLER I. Microbial transformations of mercury: Potentials, challenges, and achievements in controlling mercury toxicity in the environment [J]. Advances in Applied Microbiology, 2005, 57: 1-52. [109] FREEDMAN Z, ZHU C S, BARKAY T. Mercury resistance and mercuric reductase activities and expression among chemotrophic thermophilic Aquificae [J]. Applied and Environmental Microbiology, 2012, 78(18): 6568-6575. doi: 10.1128/AEM.01060-12 [110] BOYD E S, BARKAY T. The mercury resistance operon: From an origin in a geothermal environment to an efficient detoxification machine [J]. Frontiers in Microbiology, 2012, 3: 349. [111] MATHEMA V B, THAKURI B C, SILLANPÄÄ M. Bacterial mer operon-mediated detoxification of mercurial compounds: A short review [J]. Archives of Microbiology, 2011, 193(12): 837-844. doi: 10.1007/s00203-011-0751-4 [112] PARKS J M, GUO H, MOMANY C, et al. Mechanism of Hg-C protonolysis in the organomercurial lyase MerB [J]. Journal of the American Chemical Society, 2009, 131(37): 13278-13285. doi: 10.1021/ja9016123 [113] MELNICK J G, PARKIN G. Cleaving mercury-alkyl bonds: A functional model for mercury detoxification by MerB [J]. Science, 2007, 317(5835): 225-227. doi: 10.1126/science.1144314 [114] BARKAY T, MILLER S M, SUMMERS A O. Bacterial mercury resistance from atoms to ecosystems [J]. FEMS Microbiology Reviews, 2003, 27(2/3): 355-384. [115] BYSTROM E. Assessment of mercury methylation and demethylation with focus on chemical speciation and biological processes[D]. Dissertation, Georgia Institute of Technology, 2008. [116] LAFRANCE-VANASSE J, LEFEBVRE M, di LELLO P, et al. Crystal structures of the organomercurial lyase merb in its free and mercury-bound forms: Insights into the mechanism of methylmercury degradation [J]. Journal of Biological Chemistry, 2009, 284(2): 938-944. doi: 10.1074/jbc.M807143200 [117] LELLO P D, BENISON G C, VALAFAR H, et al. NMR structural studies reveal a novel protein fold for MerB, the organomercurial lyase involved in the bacterial mercury resistance system [J]. Biochemistry, 2004, 43(26): 8322-8332. doi: 10.1021/bi049669z [118] BENISON G C, LELLO P D, SHOKES J E, et al. A stable mercury-containing complex of the organomercurial lyase MerB: Catalysis, product release, and direct transfer to MerA [J]. Biochemistry, 2004, 43(26): 8333-8345. doi: 10.1021/bi049662h [119] PITTS K E, SUMMERS A O. The roles of thiols in the bacterial organomercurial lyase (MerB) [J]. Biochemistry, 2002, 41(32): 10287-10296. doi: 10.1021/bi0259148 [120] MAGED M, EL HOSSEINY A, SAADELDIN M K, et al. Thermal stability of a mercuric reductase from the red sea Atlantis II hot brine environment as analyzed by site-directed mutagenesis [J]. Applied and Environmental Microbiology, 2019, 85(3): e02387-e02318. [121] MØLLER A K, BARKAY T, HANSEN M A, et al. Mercuric reductase genes (merA) and mercury resistance plasmids in High Arctic snow, freshwater and sea-ice brine [J]. FEMS Microbiology Ecology, 2014, 87(1): 52-63. doi: 10.1111/1574-6941.12189 [122] SANZ-SÁEZ I, PEREIRA-GARCÍA C, BRAVO A G, et al. Prevalence of heterotrophic methylmercury detoxifying bacteria across oceanic regions [J]. Environmental Science & Technology, 2022, 56(6): 3452-3461. [123] FAGANELI J, HINES M E, COVELLI S, et al. Mercury in lagoons: An overview of the importance of the link between geochemistry and biology [J]. Estuarine, Coastal and Shelf Science, 2012, 113: 126-132. doi: 10.1016/j.ecss.2012.08.021 [124] HINES M E, FAGANELI J, ADATTO I, et al. Microbial mercury transformations in marine, estuarine and freshwater sediment downstream of the Idrija Mercury Mine, Slovenia [J]. Applied Geochemistry, 2006, 21(11): 1924-1939. doi: 10.1016/j.apgeochem.2006.08.008 [125] MARVIN-DIPASQUALE M, AGEE J, MCGOWAN C, et al. Methyl-mercury degradation pathways: A comparison among three mercury-impacted ecosystems [J]. Environmental Science & Technology, 2000, 34(23): 4908-4916. [126] OREMLAND R S, CULBERTSON C W, WINFREY M R. Methylmercury decomposition in sediments and bacterial cultures: Involvement of methanogens and sulfate reducers in oxidative demethylation [J]. Applied and Environmental Microbiology, 1991, 57(1): 130-137. doi: 10.1128/aem.57.1.130-137.1991 [127] LEE C S, FISHER N S. Methylmercury uptake by diverse marine phytoplankton [J]. Limnology and Oceanography, 2016, 61(5): 1626-1639. doi: 10.1002/lno.10318 [128] TADA Y Y, MARUMOTO K. Uptake of methylmercury by marine microalgae and its bioaccumulation in them [J]. Journal of Oceanography, 2020, 76(1): 63-70. doi: 10.1007/s10872-019-00525-6 [129] DENG G F, ZHANG T W, YANG L M, et al. Studies of biouptake and transformation of mercury by a typical unicellular diatom Phaeodactylum tricornutum [J]. Chinese Science Bulletin, 2013, 58(2): 256-265. doi: 10.1007/s11434-012-5514-3 [130] KRITEE K, MOTTA L C, BLUM J D, et al. Photomicrobial visible light-induced magnetic mass independent fractionation of mercury in a marine microalga [J]. ACS Earth and Space Chemistry, 2018, 2(5): 432-440. doi: 10.1021/acsearthspacechem.7b00056 [131] BRAVO A G, le FAUCHEUR S, MONPERRUS M, et al. Species-specific isotope tracers to study the accumulation and biotransformation of mixtures of inorganic and methyl mercury by the microalga Chlamydomonas reinhardtii [J]. Environmental Pollution, 2014, 192: 212-215. doi: 10.1016/j.envpol.2014.05.013 [132] LI Y, LI D, SONG B B, et al. The potential of mercury methylation and demethylation by 15 species of marine microalgae [J]. Water Research, 2022, 215: 118266. doi: 10.1016/j.watres.2022.118266 [133] KRISHNAMURTHY S. Biomethylation and environmental transport of metals. [J]. Journal of Chemical Education, 1992, 69(5): 347-350. doi: 10.1021/ed069p347 [134] WEBER J H. Review of possible paths for abiotic methylation of mercury(II) in the aquatic environment [J]. Chemosphere, 1993, 26(11): 2063-2077. doi: 10.1016/0045-6535(93)90032-Z [135] COSSART T, GARCIA-CALLEJA J, WORMS I A M, et al. Species-specific isotope tracking of mercury uptake and transformations by pico-nanoplankton in an eutrophic lake [J]. Environmental Pollution, 2021, 288: 117771. doi: 10.1016/j.envpol.2021.117771 [136] CERRATI G, BERNHARD M, WEBER J H. Model reactions for abiotic mercury(Ⅱ) methylation: Kinetics of methylation of mercury(Ⅱ) by mono-, di-, and tri-methyltin in seawater [J]. Applied Organometallic Chemistry, 1992, 6(7): 587-595. doi: 10.1002/aoc.590060705 [137] DESIMONE R E, PENLEY M W, CHARBONNEAU L, et al. The kinetics and mechanism of cobalamin-dependent methyl and ethyl transfer to mercuric ion [J]. Biochimica et Biophysica Acta (BBA) - General Subjects, 1973, 304(3): 851-863. doi: 10.1016/0304-4165(73)90232-8 [138] WEBER J H, REISINGER K, STOEPPLER M. Methylation of mercury (Ⅱ) by fulvic acid [J]. Environmental Technology Letters, 1985, 6(1–11): 203-208. [139] WANG K, LIU G, CAI Y. Possible pathways for mercury methylation in oxic marine waters [J]. Critical Reviews in Environmental Science and Technology, 2022, 52(22): 3997-4015. doi: 10.1080/10643389.2021.2008753 [140] JIMÉNEZ-MORENO M, PERROT V, EPOV V N, et al. Chemical kinetic isotope fractionation of mercury during abiotic methylation of Hg(II) by methylcobalamin in aqueous chloride media [J]. Chemical Geology, 2013, 336: 26-36. doi: 10.1016/j.chemgeo.2012.08.029 [141] CHEN B W, WANG T, YIN Y G, et al. Methylation of inorganic mercury by methylcobalamin in aquatic systems [J]. Applied Organometallic Chemistry, 2007, 21(6): 462-467. doi: 10.1002/aoc.1221 [142] FALTER R. Experimental study on the unintentional abiotic methylation of inorganic mercury during analysis: Part 1: Localisation of the compounds effecting the abiotic mercury methylation [J]. Chemosphere, 1999, 39(7): 1051-1073. doi: 10.1016/S0045-6535(99)00178-2 [143] KANZLER C R, LIAN P, TRAINER E L, et al. Emerging investigator series: Methylmercury speciation and dimethylmercury production in sulfidic solutions [J]. Environmental Science-Processes & Impacts, 2018, 20(4): 584-594. [144] JONSSON S, MAZRUI N M, MASON R P. Dimethylmercury formation mediated by inorganic and organic reduced sulfur surfaces [J]. Scientific Reports, 2016, 6: 27958. doi: 10.1038/srep27958 [145] CRAIG P J, BARTLETT P D. Role of hydrogen-sulfide in environmental transport of mercury [J]. Nature, 1978, 275(5681): 635-637. doi: 10.1038/275635a0 [146] KHAN M A K, WANG F Y. Chemical demethylation of methylmercury by selenoamino acids [J]. Chemical Research in Toxicology, 2010, 23(7): 1202-1206. doi: 10.1021/tx100080s [147] ASADUZZAMAN A M, KHAN M A, SCHRECKENBACH G, et al. Computational studies of structural, electronic, spectroscopic, and thermodynamic properties of methylmercury-amino acid complexes and their Se analogues [J]. Inorganic Chemistry, 2010, 49(3): 870-878. doi: 10.1021/ic900827m [148] LIAN P, MOU Z Y, COOPER C J, et al. Mechanistic investigation of dimethylmercury formation mediated by a sulfide mineral surface [J]. The Journal of Physical Chemistry. A, 2021, 125(24): 5397-5405. doi: 10.1021/acs.jpca.1c04014 [149] NI B, KRAMER J R, BELL R A, et al. Protonolysis of the Hg-C bond of chloromethylmercury and dimethylmercury. A DFT and QTAIM study [J]. The Journal of Physical Chemistry. A, 2006, 110(30): 9451-9458. doi: 10.1021/jp061852+ [150] WANG X, WANG W X. Selenium induces the demethylation of mercury in marine fish [J]. Environmental Pollution, 2017, 231: 1543-1551. doi: 10.1016/j.envpol.2017.09.014 [151] KORBAS M, O'DONOGHUE J L, WATSON G E, et al. The chemical nature of mercury in human brain following poisoning or environmental exposure [J]. ACS Chemical Neuroscience, 2010, 1(12): 810-818. doi: 10.1021/cn1000765 期刊类型引用(2)
1. 李会杰,刘雨. 矿区土壤重金属污染微生物生态修复技术研究. 环境科学与管理. 2024(02): 158-161+188 .  百度学术
百度学术
2. 夏丽,李海玥,李芹,张怀东,黄建忠. 生物对汞的耐受与转化研究进展. 武汉工程大学学报. 2024(06): 644-648 .  百度学术
百度学术
其他类型引用(1)
-






 下载:
下载:

