-
2019年,抗生素耐药性被世界卫生组织列为全球健康面临的十大威胁之一[1],抗生素耐药细菌(antibiotic resistant bacteria,ARB)和耐药基因(antibiotic resistance genes,ARGs)已成为全球人类健康的巨大威胁。饮用水作为人类重要的暴露途径,其安全性备受关注。有研究表明,全球多个国家的饮用水中均可检测到较高浓度的ARGs[2]。ARGs不仅在水环境中持续存在,还可通过可移动遗传元件(mobile genetic elements,MGEs)的介导在不同细菌间进行水平基因转移[3-4]。当ARGs转移到潜在致病菌时,可能会降低抗生素的治疗效果,对人类健康构成威胁[5]。目前,已有研究提出应从ARGs的丰度、移动性和致病性等多个角度综合评估ARGs的潜在风险[6]。因此,深入解析饮用水中ARGs的丰度、潜在移动性和潜在致病性特征,全面评估ARGs的潜在风险,将为饮用水耐药性风险控制提供理论基础。
消毒工艺是确保饮用水微生物安全最有力的屏障,也是控制饮用水中ARGs传播的重要途径。目前,自来水厂主要采用单一的消毒工艺,如氯化消毒、臭氧消毒和紫外消毒等。研究表明,氯化消毒能有效降低ARGs的绝对丰度[7],但能增加其相对丰度[8]。氯化消毒还可能增加细胞膜的通透性,从而提高ARGs水平基因转移的风险[9]。臭氧消毒可去除饮用水中的大部分ARGs,但对四环素类和磺胺类ARGs的去除效果不佳[10]。与此同时,臭氧对细胞的破坏作用会导致胞内ARGs释放,进一步增加水中ARGs的丰度[11]。紫外消毒可以影响细菌间水平基因转移的发生[12],但对不同种类ARGs的去除效果不尽相同[13]。此外,季铵盐树脂消毒因其对致病菌和外排泵类ARGs的良好去除效果备受关注[14]。然而,单一的消毒工艺难以实现ARGs的完全去除,还可能增加ARGs的耐药风险,而不同消毒工艺的联合使用可以实现优势互补,为饮用水安全提供多重保障。与此同时,从潜在移动性和潜在致病性等多角度探究饮用水消毒对ARGs耐药性风险的影响有助于揭示消毒工艺对饮用水中ARGs的控制效果。因此,有必要深入探究消毒工艺的联合使用对饮用水中ARGs分布和风险的影响。
值得关注的是,饮用水消毒可显著改变细菌群落结构,不同消毒工艺对细菌群落的影响不尽相同[8]。SHI等[15]研究发现臭氧和氯化消毒后,饮用水中残留细菌群落组成不同,臭氧消毒后优势菌属为假单胞菌属、硝化螺旋菌属和脱氯单胞菌属,而氯化消毒后假单胞菌属、军团菌属、梭状芽胞杆菌属和分枝杆菌属丰度较高。目前,越来越多的研究发现饮用水中细菌群落结构变化是导致ARGs变化的重要驱动因素,特别是细菌宿主的变化[16-17]。然而,目前缺乏针对不同消毒工艺作用下细菌宿主变化影响耐药基因组及其耐药性风险的研究。因此,需探究不同消毒工艺影响耐药性风险的潜在机制,为饮用水耐药性风险的有效控制提供理论基础。
本研究构建了氯化消毒、紫外消毒、臭氧消毒和季铵盐树脂消毒及其耦合的消毒中试工程,模拟不同消毒工艺的运行并采用基于Illumina高通量测序的宏基因组学方法全面探究消毒工艺对饮用水样品中ARGs丰度、潜在移动性和潜在致病性的影响,并评估其对ARGs耐药性风险的控制效果。在此基础上,解析饮用水样品中典型ARGs的潜在宿主,揭示ARGs耐药性风险变化的潜在机制。本研究可为饮用水微生物安全保障提供理论基础和技术支撑。
-
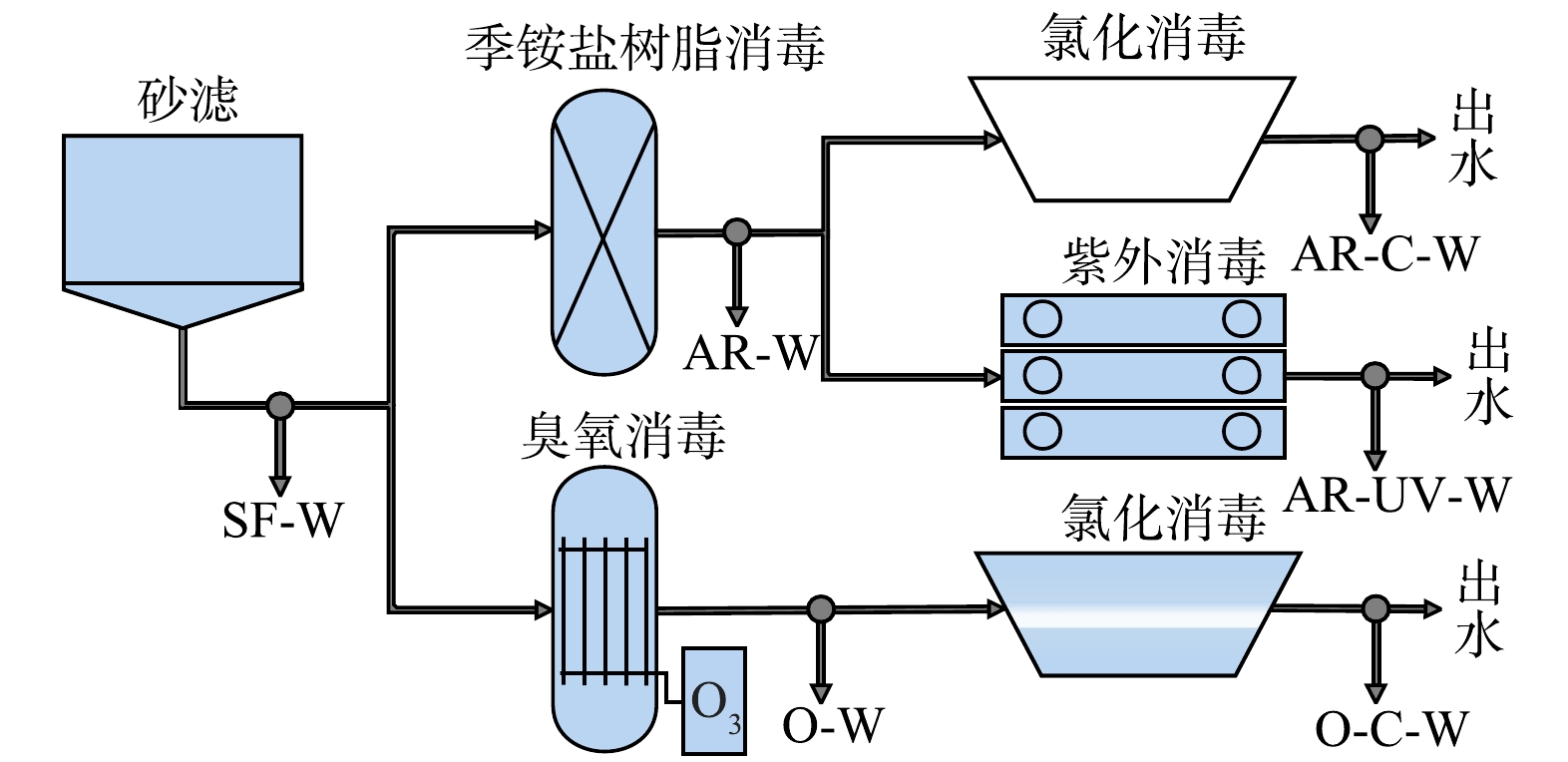
在江苏省盐城市某自来水厂构建氯化消毒、臭氧消毒、紫外消毒和季铵盐树脂消毒及其耦合的消毒中试工程(图1),每天处理水量约1 000 t,水源为长江水。其中砂滤的反冲洗频率为24 h,反冲洗时间为8 min,聚氯化铝质量浓度为200~400 mg·L−1。有效氯质量浓度为2~3 mg·L−1,反应时间为30~60 min。臭氧质量浓度为2~3 mg·L−1,反应时间为10~15 min。紫外线剂量40 mJ·cm−2。季铵盐树脂浓度为1 000~1 500 BV。采用含有0.1 μm孔径的中空丝膜的净水器分别采集砂滤出水(SF-W)、季铵盐树脂消毒出水(AR-W)、臭氧消毒出水(O-W)、臭氧-氯化消毒出水(O-C-W)、季铵盐树脂-氯化消毒出水(AR-C-W)和季铵盐树脂-紫外消毒出水(AR-UV-W),各样品的水质参数与我们之前发表的论文一致[18]。为减少水质波动的影响,在各采样点连续3次采样,每次过滤时间约为24 h,采样体积约为800 L。采样完成后,将过滤装置置于4 ℃保温箱内,4 h内转运至实验室。
-
从过滤装置中取出多层中空纤维膜,放入含有适量无菌磷酸缓冲液的离心管内,在53 kHz下超声振荡30 min,收集洗脱液。随后采用0.45 μm微孔膜过滤洗脱液,进而得到附着在膜上的细菌细胞。采用FastDNA®Spin Kit土壤试剂盒(MP Biomedicals, CA)提取样品总DNA,并检测DNA的浓度和纯度。
将提取的DNA样品送至上海美吉生物医药科技有限公司,采用Illumina HiSeq 4000平台(Illuminna Inc., San Diego, CA)进行双端测序(2×150 bp)。每个样品的原始数据大小约为10.0~14.8 Gb,平均长度为150 bp。
-
采用fastp对原始序列进行质控,使用MEGAHIT(v1.2.9)对获得的高质量reads序列进行拼接组装(默认参数)获得重叠群(contigs)。在拼接结果中筛选长度大于500 bp的重叠群进行开放性阅读框(ORF)预测。使用BLASTX将长度大于100 bp的ORF序列分别与ARGs数据库(SARG,v2.2)[19]、MGEs数据集(MGEs90.fasta)[20]和毒力因子(virulence factor genes,VFGs)核心数据集(VFDB_setA_nt.fas)[21]进行比对,使用推荐的筛选条件(e-value≤10−5,相似度≥80%和覆盖率≥70%)进行ARGs、VFGs和MGEs注释[22]。随后,使用ARGs-OAP(v2.2)估算每个样品中的细菌细胞数量(基于30组单拷贝标记基因),采用BBMap软件内部脚本pileup计算目标基因的覆盖率。为了直接表示细菌群落中目标基因的丰度水平,以各样品的细胞数量对基因拷贝数进行标准化,最终得到各基因大类/亚类的相对丰度,即目标基因拷贝数与细胞数量的比例,单位为拷贝数·细胞−1(copies·cell−1)。与此同时,以各类目标基因大类/亚类相对丰度占总相对丰度之比计算各样品中目标基因大类/亚类的百分比。
-
使用MEGAHIT将同一采样点的高质量reads序列共同组装成重叠群(默认参数),并在拼接结果中筛选长度大于500 bp的重叠群。按照上述方法依次进行ORF预测和目标基因注释,根据ARGs/MGEs/VFGs在重叠群中的存在情况判断目标基因的共存在特征。若某一重叠群中含有至少1种ARG-ORF,则为抗生素耐药重叠群(ARC)。若某一重叠群内存在2个或2个以上ARGs,则这些ARGs共存在。若某一ARC内包含至少1个MGE-ORF或VFG-ORF,则ARGs和MGEs/VFGs共存在。若某一ARC内包含至少1个MGE-ORF和VFG-ORF,则ARGs、MGEs和VFGs共存在。
-
采用Metacompare评估饮用水消毒过程中耐药性风险变化特征[23]。该方法利用CARD、ACLAME和PATRIC数据库分别注释ARGs、MGEs和人类致病菌基因序列,并根据注释结果将各样品映射到三维空间中,根据样品点s与代表最高风险的理论点h间的距离计算ARGs的风险评分。
-
采用MMseqs2(Release 14-7e284)分别对重叠群和ARC序列进行分类注释,探究饮用水中细菌群落和ARGs潜在细菌宿主的组成特征。为直接表示ARGs潜在细菌宿主的相对丰度,潜在宿主相对丰度表示为每个细胞中的目标重叠群拷贝数(拷贝数·细胞−1)。
-
使用R软件进行多样性指数计算、主坐标分析(PCoA)、多元方差分析(Adonis)和单因素方差分析。单因素方差分析中,采用Tukey HSD法进行组间多重比较,当P<0.05时,结果具有显著性。
-
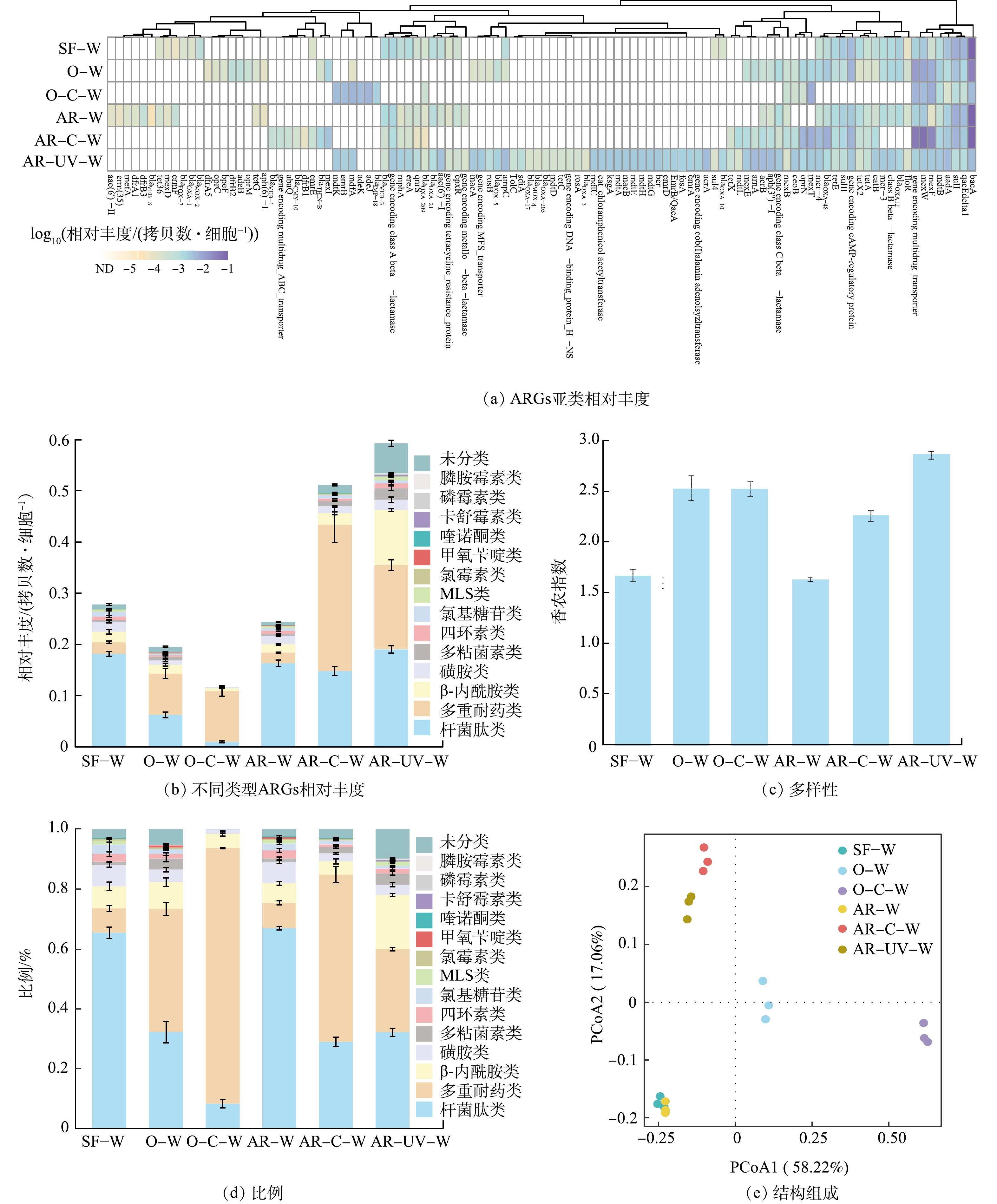
在饮用水样品中共检出15大类108亚类的ARGs,其中多重耐药类ARGs的亚类种类最丰富(图2(a)和图2(b))。季铵盐树脂消毒对ARGs多样性的影响较小,而臭氧消毒、臭氧-氯化消毒、季铵盐树脂-氯化消毒和季铵盐树脂-紫外消毒均可显著增加ARGs的多样性(P<0.05)。值得注意的是,季铵盐树脂-紫外消毒出水样品中ARGs的多样性最高(图2(c))。与此同时,饮用水样品中主要的ARGs类型为多重耐药类和杆菌肽类,这与其他研究[24]中ARGs的检测结果一致。臭氧消毒、臭氧-氯化消毒、季铵盐树脂-氯化消毒和季铵盐树脂-紫外消毒均显著提高了多重耐药类ARGs的比例(P<0.05),特别是臭氧-氯化消毒使多重耐药类ARGs的比例提升至(85.35±0.18)%(图2(d))。如图2(e)所示,采用PCoA探究ARGs组成的差异,结果表明,不同消毒处理工艺出水样品中ARGs的组成存在显著差异(Adonis:R2 = 0.961,P<0.01)。季铵盐树脂消毒出水样品和砂滤出水样品中ARGs的组成较为相似,但臭氧-氯化消毒、季铵盐树脂-氯化消毒和季铵盐树脂-紫外消毒明显改变了ARGs的组成,进一步证实了消毒工艺对ARGs的选择效应存在差异[25]。
在消毒工艺作用下,ARGs的相对丰度呈现明显差异(图2(a)和图2(b))。臭氧-氯化消毒显著降低了ARGs的总相对丰度(P<0.05),而季铵盐树脂-紫外消毒和季铵盐树脂-氯化消毒则显著增加了ARGs的总相对丰度(P<0.05),其从消毒前的(0.28±0.011) 拷贝数·细胞−1分别升高至(0.59±0.030) 拷贝数·细胞−1和(0.51±0.036) 拷贝数·细胞−1。这一结果与基于read水平的ARGs丰度结果一致,即紫外消毒和氯化消毒会进一步富集ARGs,提高其相对丰度[8]。与季铵盐树脂消毒出水样品相比,紫外和氯化消毒均显著增加了多重耐药类ARGs的相对丰度(P<0.05),特别是季铵盐树脂-氯化消毒出水样品中多重耐药类ARGs的相对丰度是砂滤出水的12.71倍,其中mexF和mexW等ARGs贡献较大。已有较多研究表明,氯化消毒和紫外消毒能显著增加多重耐药类ARGs的相对丰度[17,26]。臭氧消毒和臭氧-氯化消毒处理后,多重耐药类ARGs的相对丰度显著增加(P<0.05),这可能是因为多重耐药菌对低剂量臭氧消毒具有较强的耐受性[25]。此外,臭氧消毒显著降低了杆菌肽类ARGs的相对丰度(P<0.05),而随后的氯化消毒进一步降低其丰度。有研究表明,杆菌肽类ARGs的同源基因存在于153个细菌属中,这表明杆菌肽类ARGs可能是细菌固有的基因[22],在环境压力下容易与细菌宿主一同被去除。
-
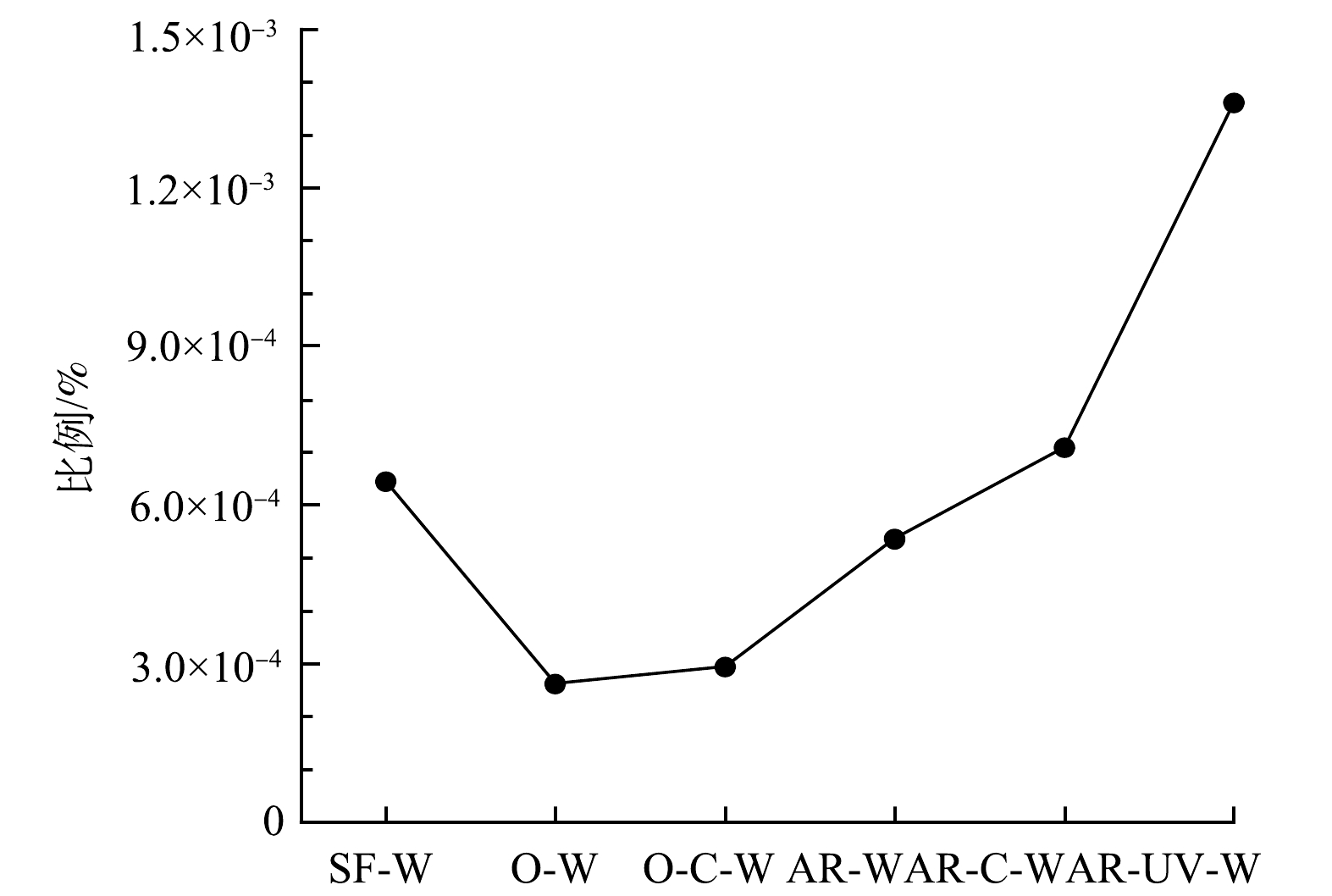
为探究不同消毒工艺对ARGs共存在特征的影响,本研究采用基于高通量测序的组装和功能注释方法,在饮用水样品中筛选出同时携带2个及2个以上ARGs的ARCs,并分析其比例变化和ARGs的排布特征。如图3所示,各组样品中携带多个ARGs的ARCs的比例不同。臭氧消毒明显降低了ARGs共存在的比例,但随后的氯化消毒却小幅增加了这一比例,这可能是因为氯化消毒导致非耐氯菌胞内ARGs释放到水环境中,促进了环境中ARGs的水平基因转移[9]。值得注意的是,季铵盐树脂对ARGs共存在的影响较小,但在季铵盐树脂-紫外消毒出水样品中ARGs共存在的比例最高(0.001 4%)。此外,在饮用水样品中共发现21种ARGs共存在排布模式。如表1所示,在相对丰度大于0.001 拷贝数·细胞−1的同时携带2个及2个以上ARGs的ARCs中,携带多重耐药类和磺胺类ARGs的ARCs以及携带多种多重耐药类ARGs的ARCs的相对丰度最高,这可能是由于环境中抗生素的共同选择作用[24,27]。
-
图4显示了饮用水样品中MGEs的组成和相对丰度。饮用水样品中主要的MGEs为转座酶基因、重组酶基因和整合酶基因。臭氧消毒后,MGEs的总相对丰度最低,为(7.43±0.15) 拷贝数·细胞−1,随后的氯化消毒导致MGEs的相对丰度略有上升,但仍低于砂滤出水样品。这与之前的研究相符,表明氯化消毒对MGEs有富集作用[28]。与此同时,季铵盐树脂消毒降低了MGEs的相对丰度,但随后的氯化消毒和紫外消毒显著提升MGEs的总相对丰度至(13.12±0.25) 拷贝数·细胞−1和(14.45±0.45) 拷贝数·细胞−1(P<0.05)。其中,转座酶基因相对丰度的增加对MGEs总丰度的提升贡献较大。
通过分析ARGs与MGEs的共存在特征,进一步评估了饮用水消毒对ARGs移动性的影响(图4(b)和表2)。臭氧消毒后,同时携带ARGs与MGEs的重叠群占总重叠群的比例最低(0.000 4%),但随后的氯化消毒使其比例略有增加。季铵盐树脂消毒促进了ARGs与MGEs的共存在,且这种促进作用在后续的紫外消毒中进一步加强,导致季铵盐树脂-紫外消毒出水样品中同时携带ARGs与MGEs的重叠群的比例最高(0.001 1%)。此外,在饮用水样品中共发现31种ARGs与MGEs的共存在关系,表2显示了相对丰度大于0.001 拷贝数·细胞−1的同时携带ARGs与MGEs的重叠群。与ARGs存在线性排列的MGEs主要有转座酶、重组酶和整合酶,其中转座酶基因与多种ARGs在所有样品中均共存,表明转座酶基因可能在ARGs的传播中具有重要作用。此外,研究还发现,同时携带多重耐药类/磺胺类ARGs与MGEs的重叠群的相对丰度最高,这与KE等[29]的研究结果一致,表明多重耐药类ARGs与磺胺类ARGs可能具有较强的潜在移动性。
-
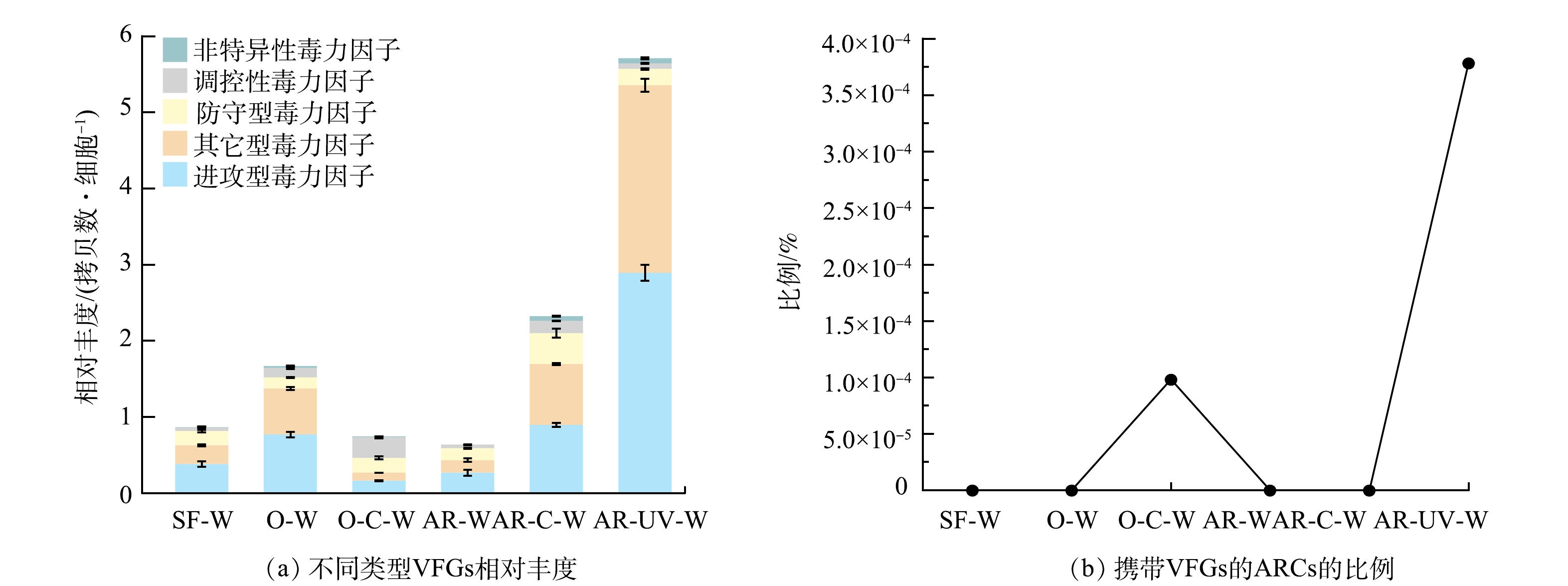
在饮用水样品中共检出5大类17亚类VFGs,其中进攻型和其它型VFGs为主要的VFGs。图5(a)反映了不同消毒工艺处理过程中VFGs相对丰度的变化趋势。臭氧-氯化消毒出水样品中VFGs的总相对丰度与砂滤出水相比无显著差异,但季铵盐树脂-氯化消毒和季铵盐树脂-紫外消毒显著提高了VFGs的总相对丰度(P<0.05)。值得注意的是,季铵盐树脂-紫外消毒对VFGs的提升作用最显著,其将VFGs的总相对丰度显著提高至(5.71±0.17) 拷贝数·细胞−1 (P<0.05),其中进攻型VFGs和其它型VFGs的相对丰度也显著提高(P<0.05),对VFGs总丰度的提升贡献最大。LIANG等[30]的研究指出高丰度的进攻型VFGs可以增强宿主的感染能力,而高丰度的防守型VFGs可以增强宿主免疫系统的耐受能力,这表明饮用水消毒对含有VFGs的宿主细菌的感染能力和免疫系统耐受能力有潜在影响。
如图5(b)所示,不同消毒工艺作用对ARGs和VFGs的共存在特征的影响具有较大差异,仅在臭氧-氯化消毒出水样品和季铵盐树脂-紫外消毒出水样品中发现ARGs与VFGs的共存在,且季铵盐树脂-紫外消毒极大地促进了ARGs与VFGs的共存在。有研究将这些同时携带ARGs与VFGs的菌株视为具有潜在致病性的抗生素耐药细菌[30]。此外,在饮用水样品中共发现了6种ARGs与VFGs的共存在关系,其中在大部分样品中发现VFGs与多重耐药类ARGs的共存在(表3)。这一结果与之前的研究结果一致,发现进攻型VFGs与多重耐药类ARGs的基因组合是主要的共存模式[30]。在季铵盐树脂-紫外消毒样品中发现多个ARGs和多个VFGs的基因线性组合,例如某些重叠群上同时含有2个多重耐药类ARGs、3个进攻型VFGs和1个其它型VFGs(表3)。值得关注的是,臭氧-氯化消毒出水样品中同时携带多重耐药类ARGs和防守型VFGs的重叠群上还存在MGEs(表3),这可能极大增加了ARGs和VFGs的水平转移风险。
-
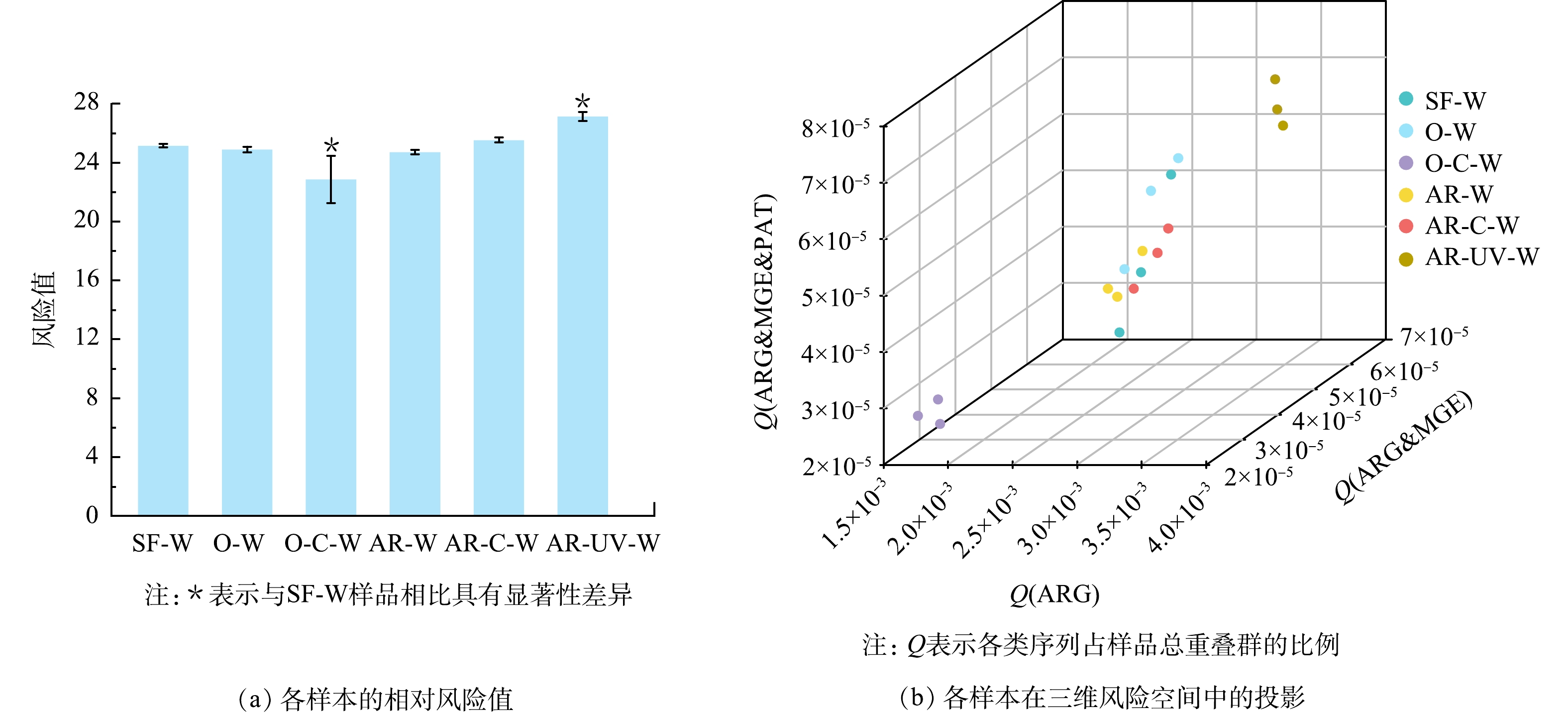
ARGs的潜在风险主要与其水平转移能力及其与致病菌的关联密切相关。因此,对ARGs潜在风险的评估需要综合考虑ARGs的相对丰度、可移动性以及在致病菌内的存在情况。图6(a)为采用Metacompare计算的饮用水样品中ARGs的潜在风险值。结果表明,与砂滤出水样品相比,臭氧消毒和季铵盐树脂消毒对ARGs的相对风险影响较小,臭氧-氯化消毒显著降低了ARGs的相对风险值(P<0.05)。然而,先前有研究发现氯化消毒会导致医院废水中ARGs的风险值升高[31],这表明氯化消毒对ARGs潜在风险的影响较为复杂,可能与氯的浓度和消毒时间有关。与砂滤出水样品相比,季铵盐树脂-氯化消毒和季铵盐树脂-紫外消毒均增加了ARGs的潜在风险,其中季铵盐树脂-紫外消毒的影响具有显著性(P<0.05),且出水中ARGs的相对风险值最高,为27.13±0.31。如图6(b)所示,各样品在三维风险空间中的投影也反映了这一结果,季铵盐树脂-紫外消毒出水样品中ARGs相关序列、ARGs潜在移动性相关序列以及致病菌携带的ARGs相关序列的比例均达到最大值,与之前ARGs相对丰度、潜在移动性和潜在致病性的结果一致。
-
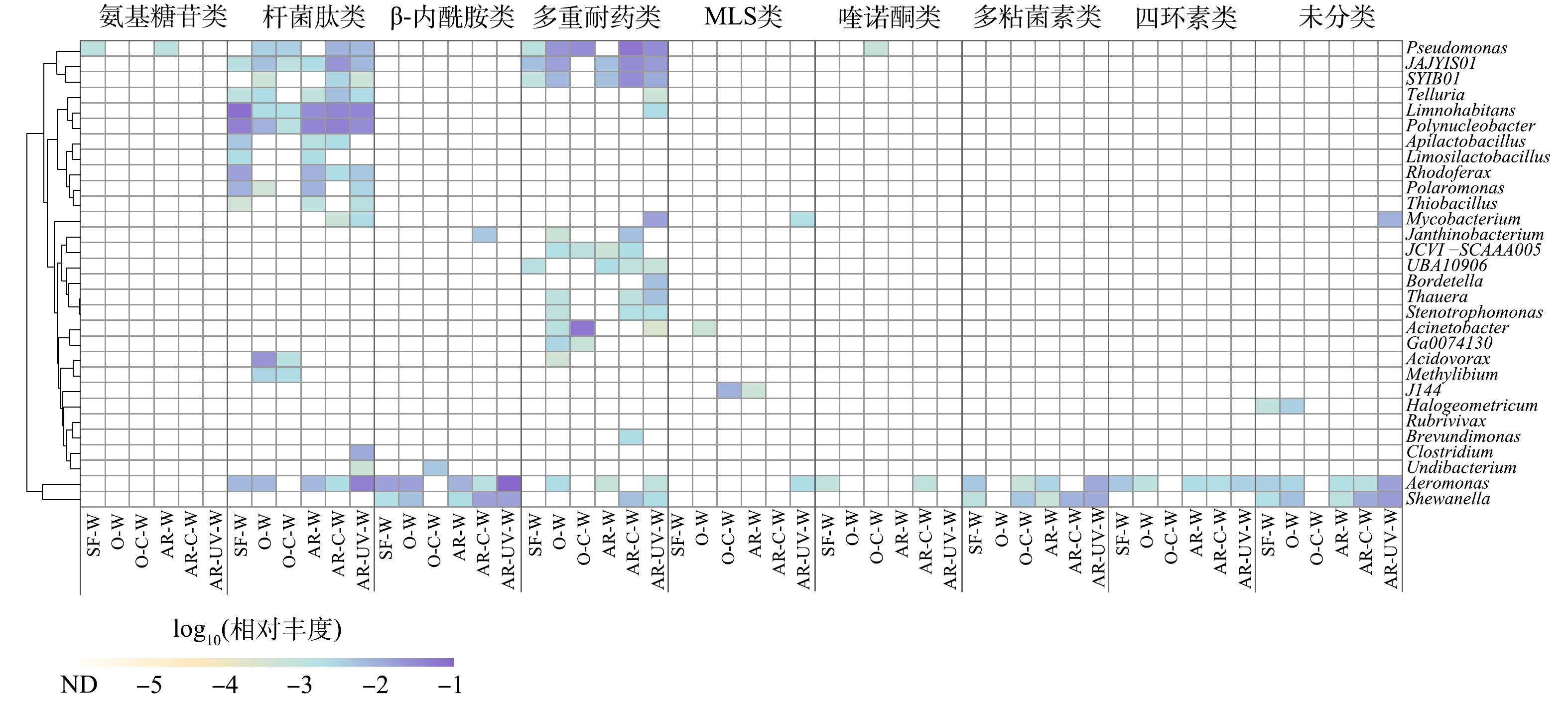
目前,已有研究表明细菌群落结构是影响ARGs分布的重要因素,各类消毒工艺能够改变细菌群落结构组成和多样性[17-18]。值得关注的是,细菌宿主是饮用水中ARGs变化的重要驱动因素[17],探究消毒对ARGs潜在细菌宿主组成的和丰度的影响对于揭示饮用水消毒影响细菌耐药性的机制具有重要意义。因此,本研究对饮用水消毒过程中ARGs的潜在宿主进行了属水平的分类注释,以揭示ARGs宿主在不同消毒过程中的变化。如图7所示,气单胞菌属(Aeromonas)和假单胞菌属(Pseudomonas)等相对丰度最高的30个菌属携带氨基糖苷类、杆菌肽类、β-内酰胺类、多重耐药类、MLS类、喹诺酮类、多粘菌素类、四环素类以及未分类ARGs,是其潜在宿主。消毒改变了ARGs潜在宿主的组成和丰度,尤其是多重耐药类和杆菌肽类ARGs的细菌宿主。在季铵盐树脂-氯化消毒和季铵盐树脂-紫外消毒出水样品中,多重耐药类ARGs的主要宿主是假单胞菌属,而臭氧-氯化消毒后的主要宿主为不动杆菌属(Acinetobacter)。值得注意的是,季铵盐树脂-氯化消毒和季铵盐树脂-紫外消毒均大幅提高了多重耐药类ARGs潜在宿主假单胞菌属的相对丰度,特别是季铵盐树脂-氯化消毒出水样品中假单胞菌属的相对丰度为砂滤出水的15.42倍。假单胞菌属对氯暴露和紫外辐射具有较强的耐受性[25,32]。SHI等[15]的研究也发现假单胞菌属是氯化消毒和臭氧消毒后的主要残留菌属,其在水环境中的传播值得关注。已有研究发现细菌对消毒剂和抗生素的交叉耐药性可导致细菌更易在选择压力下存活[18],这解释了季铵盐树脂-氯化消毒和季铵盐树脂-紫外消毒对假单胞菌的富集作用。KE等[29]的研究指出这种交叉耐药性可能与外排泵有关,可能导致细菌产生多重耐药性。已有大量文献报道假单胞菌携带多种ARGs,是促进ARGs增殖和传播的最重要菌属之一[2,33],消毒对其的富集作用可能导致多重耐药类ARGs相对丰度的提高。与此同时,季铵盐树脂-氯化消毒后,杆菌肽类ARGs的主要宿主是多核杆菌属(Polynucleobacter)和栖湖菌属(Limnohabitans),而季铵盐树脂-紫外消毒处理后,杆菌肽类ARGs主要宿主是气单胞菌属、栖湖菌属和多核杆菌属。臭氧消毒后,杆菌肽类ARGs的主要宿主为食酸菌属(Acidovorax),然而,在随后的氯化消毒过程中,这些ARGs的主要潜在宿主为假单胞菌属。值得注意的是,臭氧-氯化消毒后杆菌肽类ARGs及其细菌潜在宿主的总相对丰度均降至最低,与先前的研究结果一致[34],表明氯化消毒可能通过对细菌宿主的去除实现对杆菌肽类ARGs的去除。总体而言,明确关键细菌宿主组成可对ARGs风险控制具有重要意义。
与此同时,水平基因转移被认为是细菌获得抗生素耐药性的重要分子机制,大量研究发现MGEs可以促进ARGs在水环境中的传播。目前,已有研究报道饮用水消毒出水中多重耐药类ARGs(mexH)和重组酶基因(recR)的共存在关系[18]。本研究发现季铵盐树脂-紫外消毒出水样品中同时携带ARGs与MGEs的重叠群的比例较其他消毒工艺出水高,表明此种消毒工艺对ARGs在不同细菌宿主间的水平转移具有较强的促进作用。此外,多个ARGs与MGEs共存于一条重叠群,表明MGEs有可能导致共抗性或多重抗性的形成[35]。值得注意的是,转座酶基因是与ARGs具有共存在关系的主要MGEs类型。已有研究表明,在多种环境介质中均检测到转座酶基因tnpA-04、tnpA-07和tP614等,其可能对ARGs在细菌间的转移和获得具有重要作用[18]。本研究发现转座酶基因与多重耐药类和磺胺类ARGs共存在,表明多重耐药类和磺胺类ARGs在环境中的传播潜力更大,这与CHEN等[36]的研究结果一致,在河流和湖泊交汇的底泥中转座酶基因(IS91和tnpA)经常携带多重耐药类与磺胺类ARGs,MGEs(ISCau1和tnpAcp2等)对底泥中ARGs分布具有重要作用。
-
1)本研究采集的饮用水样品中多重耐药类和杆菌肽类ARGs的相对丰度较高。季铵盐树脂-氯化消毒和季铵盐树脂-紫外消毒显著增加了饮用水样品中ARGs、MGEs和VFGs的总相对丰度,增加了ARGs的潜在风险值。季铵盐树脂-紫外消毒还促进了ARGs与MGEs和VFGs的共存潜力。
2)消毒改变了饮用水样品中ARGs潜在宿主的组成和相对丰度。季铵盐树脂-氯化消毒和季铵盐树脂-紫外消毒提高了多重耐药类ARGs宿主假单胞菌属的相对丰度,可能导致多重耐药性。
3)消毒导致耐药性风险变化的潜在机制还需深入研究,可利用第三代测序技术的读长优势进一步阐明消毒影响细菌耐药性的分子生态学机理。
饮用水消毒工艺对抗生素耐药性风险的影响及其机理
Influence and mechanism of drinking water disinfection strategies on antibiotic resistance risk
-
摘要: 为探究饮用水消毒工艺对耐药性风险的影响及其机制,本研究模拟了中试运行条件下不同消毒工艺的运行,采用基于高通量测序的宏基因组学方法分析不同消毒工艺对抗生素耐药基因(ARGs)丰度、潜在移动性和潜在致病性的影响,综合评估其对ARGs风险的控制效果并解析其机制。结果表明,臭氧-氯化消毒(O-C)显著降低了ARGs的总相对丰度,季铵盐树脂-氯化消毒(AR-C)和季铵盐树脂-紫外消毒(AR-UV)则显著增加了ARGs的总相对丰度。同时,AR-C和AR-UV增加了可移动遗传元件(MGEs)的总相对丰度,促进了ARGs与MGEs的共存在。此外,这2种消毒工艺提高了毒力因子(VFGs)的总相对丰度,其中进攻型和其他型VFGs的贡献最大。AR-UV还能促进ARGs与VFGs的共存在,多重耐药类ARGs与VFGs的基因组合是主要的共存模式。总的来说,AR-C和AR-UV提高了ARGs的潜在风险。消毒改变了ARGs潜在宿主的组成和丰度,宿主改变和水平基因转移是消毒过程中ARGs变化的关键因素。Abstract: To explore the impact and mechanism of drinking water disinfection on the risk of antibiotic resistance, the operation of different disinfection processes was simulated under pilot conditions in this study, and the metagenomic approach based on high-throughput sequencing was employed to analyze the effects of different disinfection processes on the abundance, potential mobility, and potential pathogenicity of antibiotic resistance genes (ARGs). Their control effectiveness on ARGs risk was comprehensively evaluated and the underlying mechanisms were deciphered. The results indicated that ozone-chlorine disinfection(O-C) significantly reduced the total relative abundance of ARGs, while antimicrobial resin-chlorine disinfection (AR-C) and antimicrobial resin-ultraviolet disinfection (AR-UV) significantly increased the total relative abundance of ARGs. Simultaneously, AR-C and AR-UV increased the total relative abundance of mobile genetic elements (MGEs), promoting the co-occurrence of ARGs and MGEs. In addition, these two disinfection processes increased the total relative abundance of virulence factor genes (VFGs), of which the offensive and other VFGs made the most contribution. AR-UV also facilitated the co-occurrence of ARGs and VFGs, and the gene combination of multidrug resistance genes and VFGs was the main co-occurrence mode. Overall, AR-C and AR-UV increased the potential risk of ARGs. Disinfection altered the composition and abundance of ARGs potential hosts, and host changes and horizontal gene transfer were the key factors for the variation of ARGs during the disinfection process.
-
表 1 饮用水样品中共存在ARGs的排布特征
Table 1. The distribution characteristics of co-occurred ARGs in drinking water samples
样品 ARGs的排布特征 相对丰度(拷贝数·细胞−1) SF-W 多重耐药类-磺胺类 0.012 5 氨基糖胺类-β内酰胺类 0.001 5 氨基糖胺类-氨基糖胺类 0.001 4 β-内酰胺类-磺胺类 0.001 4 氨基糖胺类-大环内酯类 0.001 4 O-W 多重耐药类-磺胺类 0.003 7 多重耐药类-多重耐药类 0.001 3 O-C-W 多重耐药类-多重耐药类 0.013 9 多重耐药类-磺胺类 0.001 3 AR-W 多重耐药类-磺胺类 0.010 5 氨基糖苷类-β-内酰胺类 0.002 6 杆菌肽类-杆菌肽类 0.002 1 AR-C-W 多重耐药类-磺胺类 0.008 9 多重耐药类-多重耐药类 0.005 1 β-内酰胺类-磺胺类 0.001 7 β-内酰胺类-氨基糖苷类 0.001 6 杆菌肽类-杆菌肽类 0.001 5 氯霉素类-甲氧苄啶 0.001 1 氨基糖苷类-氨基糖苷类 0.001 0 AR-UV-W 多重耐药类-磺胺类 0.009 8 多重耐药类-多重耐药类 0.008 5 磷霉素类-卡数霉素类 0.002 1 杆菌肽类-多重耐药类 0.002 1 大环内酯类-大环内酯类 0.001 6 多重耐药类(4)-未分类 0.001 6 未分类-未分类 0.001 3 注:括号内数字代表基因数量。 表 2 饮用水样品中ARGs与MGEs的排布特征
Table 2. The distribution characteristics of ARGs and MGEs in drinking water samples
样品 ARGs与MGEs的排布特征 相对丰度(拷贝数·细胞−1) SF-W 磺胺类-转座酶 0.005 7 磺胺类-重组酶-转座酶 0.002 0 四环素类-转座酶 0.001 9 β-内酰胺类-磺胺类-转座酶 0.001 4 O-W 杆菌肽类-整合酶 0.019 8 多重耐药类-磺胺类-转座酶 0.003 7 磺胺类-转座酶 0.001 1 O-C-W 多重耐药类-转座酶(2) 0.011 1 多重耐药类(2)-重组酶-转座酶 0.010 4 多重耐药类-整合酶(2)-重组酶-转座酶 0.010 2 磺胺类-转座酶 0.002 0 AR-W 氨基糖苷类-转座酶(2) 0.003 2 氨基糖苷类-β-内酰胺类-转座酶 0.002 6 磺胺类-转座酶(2) 0.002 3 β-内酰胺类-转座酶 0.001 5 β-内酰胺类-重组酶-转座酶 0.001 4 AR-C-W 多重耐药类-转座酶 0.003 2 四环素类-转座酶 0.001 9 β-内酰磺胺类-磺胺类-转座酶 0.001 7 氨基糖苷类(2)-重组酶-转座酶 0.001 0 AR-UV-W 多粘菌素-整合酶 0.011 1 多重耐药类-磺胺类-重组酶 0.009 8 多重耐药类-转座酶(2) 0.004 1 磺胺类-重组酶-转座酶(2) 0.003 3 β-内酰磺胺类-重组酶-转座酶(2) 0.002 4 磷霉素-卡舒霉素-转座酶-整合酶 0.002 1 多重耐药类(2)-整合酶 0.001 9 多重耐药类-整合酶 0.001 7 杆菌肽类-转座酶 0.001 6 氨基糖苷类-转座酶 0.001 5 注:括号内数字代表基因数量。 表 3 饮用水样品中ARGs与VFGs的排布特征
Table 3. The distribution characteristics of ARGs and VFGs in drinking water samples
样品 ARGs与VFGs的排布特征 相对丰度(拷贝数·细胞−1) O-W 多重耐药类-防守型VFGs-转座酶 0.011 1 AR-UV-W 多重耐药类-调控型VFGs 0.001 7 未分类-多重耐药类(4)-其它型VFGs(5)-防守型VFGs(4) 0.001 6 多重耐药类(2)-进攻型VFGs(3)-其它型VFGs 0.001 5 未分类(2)-进攻型VFGs 0.001 3 多重耐药类-非特异性VFGs 0.001 3 注:括号内数字代表基因数量。 -
[1] THANGARAJU P, VENKATESAN S. WHO ten threats to global health in 2019: antimicrobial resistance[J]. Geneva:Cukurova Medical Journal, 2019, 44(3): 1150-1151. doi: 10.17826/cumj.514157 [2] MA L P, LI B, JIANG X T, et al. Catalogue of antibiotic resistome and host-tracking in drinking water deciphered by a large scale survey[J]. Microbiome, 2017, 5: 154. doi: 10.1186/s40168-017-0369-0 [3] GILLINGS M R, GAZE W H, PRUDEN A, et al. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution[J]. The ISME Journal, 2015, 9(6): 1269-1279. doi: 10.1038/ismej.2014.226 [4] STOKES H W, NESBO C L, HOLLEY M, et al. Class 1 integrons potentially predating the association with Tn402-like transposition genes are present in a sediment microbial community[J]. Journal of Bacteriology, 2006, 188(16): 5722-5730. doi: 10.1128/JB.01950-05 [5] LUO Y, XU L, RYSZ M, et al. Occurrence and transport of tetracycline, sulfonamide, quinolone, and macrolide antibiotics in the Haihe River Basin, China[J]. Environmental Science & Technology, 2011, 45(5): 1827-1833. [6] ZHANG A N, GASTON J M, DAI C L, et al. An omics-based framework for assessing the health risk of antimicrobial resistance genes[J]. Nature Communications, 2021, 12(1): 4765. doi: 10.1038/s41467-021-25096-3 [7] CHAO Y Q, MA L P, YANG Y, et al. Metagenomic analysis reveals significant changes of microbial compositions and protective functions during drinking water treatment[J]. Scientific Reports, 2013, 3: 3550. doi: 10.1038/srep03550 [8] JIA S Y, BIAN K Q, SHI P, et al. Metagenomic profiling of antibiotic resistance genes and their associations with bacterial community during multiple disinfection regimes in a full-scale drinking water treatment plant[J]. Water Research, 2020, 176: 115721. doi: 10.1016/j.watres.2020.115721 [9] ZHANG S, WANG Y, LU J, et al. Chlorine disinfection facilitates natural transformation through ROS-mediated oxidative stress[J]. Isme Journal, 2021, 15(10): 2969-2985. doi: 10.1038/s41396-021-00980-4 [10] HU Y R, JIANG L, ZHANG T Y, et al. Occurrence and removal of sulfonamide antibiotics and antibiotic resistance genes in conventional and advanced drinking water treatment processes[J]. Journal of Hazardous Materials, 2018, 360: 364-372. doi: 10.1016/j.jhazmat.2018.08.012 [11] WANG J, SHA X N, CHEN X F, et al. Removal and distribution of antibiotics and resistance genes in conventional and advanced drinking water treatment processes[J]. Journal of Water Process Engineering, 2022, 50: 103217. doi: 10.1016/j.jwpe.2022.103217 [12] 林文芳. 饮用水中细菌抗生素抗性的共选择、水平基因转移及稳定性研究[D]. 北京: 中国科学院大学, 2016. [13] 张明露, 徐梦瑶, 王礼, 等. 紫外消毒对管网多相界面中抗性基因的影响[J]. 给水排水, 2018, 44(5): 42-46. doi: 10.3969/j.issn.1002-8471.2018.05.011 [14] 常芳瑜. 季铵盐树脂对饮用水中致病菌以及抗生素抗性基因的控制技术原理[D]. 南京: 南京大学, 2018. [15] SHI Q, CHEN Z, YAN H, et al. Identification of significant live bacterial community shifts in different reclaimed waters during ozone and chlorine disinfection[J]. Science of the Total Environment, 2023, 896: 165199. doi: 10.1016/j.scitotenv.2023.165199 [16] XU L K, CAMPOS L C, CANALES M, et al. Drinking water biofiltration: Behaviour of antibiotic resistance genes and the association with bacterial community[J]. Water Research, 2020, 182: 115954. doi: 10.1016/j.watres.2020.115954 [17] JIA S Y, SHI P, HU Q, et al. Bacterial community shift drives antibiotic resistance promotion during drinking wter chlorination[J]. Environmental Science & Technology, 2015, 49(20): 12271-12279. [18] ZHANG H C, CHANG F Y, SHI P, et al. Antibiotic resistome alteration by different disinfection strategies in a full-scale drinking water treatment plant deciphered by metagenomic assembly[J]. Environmental Science & Technology, 2019, 53(4): 2141-2150. [19] YIN X L, JIANG X T, CHAI B L, et al. ARGs-OAP v2.0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes[J]. Bioinformatics, 2018, 34(13): 2263-2270. doi: 10.1093/bioinformatics/bty053 [20] WU D, JIN L, XIE J W, et al. Inhalable antibiotic resistomes emitted from hospitals: metagenomic insights into bacterial hosts, clinical relevance, and environmental risks[J]. Microbiome, 2022, 10(1): 19. doi: 10.1186/s40168-021-01197-5 [21] CHEN L H, YANG J, YU J, et al. VFDB: a reference database for bacterial virulence factors[J]. Nucleic Acids Research, 2005, 33(Database issue): D325-D328. [22] HU Y F, YANG X, QIN J J, et al. Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota[J]. Nature Communications, 2013, 4: 2151. doi: 10.1038/ncomms3151 [23] OH M, PRUDEN A, CHEN C Q, et al. MetaCompare: a computational pipeline for prioritizing environmental resistome risk[J]. FEMS Microbiology Ecology, 2018, 94(7): fiy079. [24] ZHAO Q Q, HE H, GAO K, et al. Fate, mobility, and pathogenicity of drinking water treatment plant resistomes deciZHIered by metagenomic assembly and network analyses[J]. Science of the Total Environment, 2022, 804: 150095. doi: 10.1016/j.scitotenv.2021.150095 [25] ZHENG J, SU C, ZHOU J W, et al. Effects and mechanisms of ultraviolet, chlorination, and ozone disinfection on antibiotic resistance genes in secondary effluents of municipal wastewater treatment plants[J]. Chemical Engineering Journal, 2017, 317: 309-316. doi: 10.1016/j.cej.2017.02.076 [26] WOOD D E, LU J, LANGMEAD B. Improved metagenomic analysis with Kraken 2[J]. Genome biology, 2019, 20(1): 257. doi: 10.1186/s13059-019-1891-0 [27] JU F, LI B, MA L P, et al. Antibiotic resistance genes and human bacterial pathogens: Co-occurrence, removal, and enrichment in municipal sewage sludge digesters[J]. Water Research, 2016, 91: 1-10. doi: 10.1016/j.watres.2015.11.071 [28] SHI P, JIA S, ZHANG X X, et al. Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water[J]. Water Research, 2013, 47: 111-120. doi: 10.1016/j.watres.2012.09.046 [29] KE Y C, SUN W J, JING Z B, et al. Seasonal variations of microbial community and antibiotic resistome in a suburb drinking water distribution system in a northern Chinese city[J]. Journal of Environmental Sciences, 2023, 127: 714-725. doi: 10.1016/j.jes.2022.07.001 [30] LIANG J S, MAO G N, YIN X L, et al. Identification and quantification of bacterial genomes carrying antibiotic resistance genes and virulence factor genes for aquatic microbiological risk assessment[J]. Water Research, 2020, 168: 115160. doi: 10.1016/j.watres.2019.115160 [31] ROLBIECKI D, PAUKSZTO L, KRAWCZYK K, et al. Chlorine disinfection modifies the microbiome, resistome and mobilome of hospital wastewater: A nanopore long-read metagenomic approach[J]. Journal of Hazardous Materials, 2023, 459: 132298. doi: 10.1016/j.jhazmat.2023.132298 [32] HU Q, ZHANG X X, JIA S Y, et al. Metagenomic insights into ultraviolet disinfection effects on antibiotic resistome in biologically treated wastewater[J]. Water Research, 2016, 101: 309-317. doi: 10.1016/j.watres.2016.05.092 [33] SU H C, LIU Y S, PAN C G, et al. Persistence of antibiotic resistance genes and bacterial community changes in drinking water treatment system: From drinking water source to tap water[J]. Science of the Total Environment, 2018, 616: 453-461. [34] KE Y C, SUN W J, JING Z B, et al. Antibiotic resistome alteration along a full-scale drinking water supply system deciphered by metagenome assembly: Regulated by seasonality, mobile gene elements and antibiotic resistant gene hosts[J]. Science of the Total Environment, 2023, 862: 160887. doi: 10.1016/j.scitotenv.2022.160887 [35] HE L Y, LIU Y S, SU H C, et al. Dissemination of antibiotic resistance genes in representative broiler feedlots environments: Identification of indicator ARGs and correlations with environmental variables[J]. Environmental Science & Technology, 2014, 48(22): 13120-13129. [36] CHEN H Y, LI Y, Z SUN W C, et al. Characterization and source identification of antibiotic resistance genes in the sediments of an interconnected river-lake system[J]. Environment International, 2020, 137: 105538. doi: 10.1016/j.envint.2020.105538 -




 下载:
下载: