-
氮素是导致水体富营养化的重要营养物质之一,污水处理厂总氮排放标准为15~20 mg L−1,远高于地表水环境质量标准(GB 3838-2002)。过量的氮排入地表水会造成水体富营养化。因此,对污处理水厂的二级处理出水进行再处理尤为迫切,而潜流型人工湿地因其处理效率高和具有多重生态服务功能而在污水再处理中得到广泛应用[1]。潜流型人工湿地是通过植物、微生物、基质间的协同作用实现氮去除。有研究[2-5]表明,植物吸收和基质吸附对氮去除的贡献较小,微生物对氮的转化利用才是主要的脱氮途径。潜流型人工湿地微生物脱氮主要依靠硝化和反硝化过程[6],介导硝化反应的细菌主要是好氧的自养微生物,而介导反硝化过程的微生物主要是厌氧的异养微生物[7],且该过程需要碳源提供电子受体。传统的人工湿地中总氮去除率为40%~55%[8],很多人工湿地总氮处理效率均低于50%[9]。氮去除主要受氧气和有机碳不足的限制[10],因此,亟需采取强化措施提高人工湿地脱氮效率。LAI等[11]对曝气条件下的垂直流人工湿地的脱氮效果进行了研究并发现,在C/N由3增加到12的过程中,TN、
NO−3 、COD去除率随C/N增加而增加,但C/N过高会抑制NH+4 的去除。CHEN 等[12]的研究表明,在无曝气、碳源缺乏条件下(C/N=1.6),因NO−3 与NO−2 竞争电子受体,导致NO−2 积累;而在C/N增加到2.8时,在种植植物下TN去除率可达99%,且无NO−2 积累。曝气和C/N对低C/N污水中氮转化途径具有重要影响,然而,曝气条件下低C/N污水处理效果和微生物对不同C/N的响应仍不清楚。在植物生长旺盛期,植物根系分泌物中的可溶性糖和小分子有机酸(乙酸、草酸、琥珀酸等)可以为反硝化微生物提供部分碳源[13]。已有文献中,多采用5∶1、10∶1、15∶1等较高的C/N研究不同C/N对人工湿地污水处理效果[14-15]。然而在高C/N条件下,虽然氮去除效率较高,但也会消耗更多的可溶性氧,会限制好氧硝化过程;同时,过量的碳源也会增加二次污染的风险[16-17]。因此,只有平衡碳源和可溶性氧的需求才能实现最佳的氮去除效果,节能减耗。本研究在芦苇旺盛期开展,评价了在曝气条件下不同低C/N(0.9∶1,2∶1,4∶1)对污水的处理效果,阐明了在低C/N污水处理中氮去除的微生物机制,以期为水平潜流型人工湿地的节能运行提供参考。
全文HTML
-
供试湿地为水平潜流型人工湿地,位于济南市西区污水厂,该区域属暖温带大陆性季风气候。共设有3个湿地单元,湿地单元面积为24.05 m2 (长6.50 m,宽3.70 m),每个湿地单元设有独立的进水系统和曝气装置。水平潜流型人工湿地基质由下而上依次是粗砂、粗砾石、细砾石、细砂、粗砂、土壤种植层,各层厚度分别为0.10、0.30、0.30、0.15、0.10、0.20 m。湿地进水为污水处理厂的尾水,尾水排放执行《城镇污水处理厂污染物排放标准》(GB 18918-2002)中的一级A标准,COD、
NH+4 、TN、TP年均浓度为20.00、7.20、22.00、2.01 mg·L−1。湿地进水为连续进水,人工湿地水力负荷为0.33 m3·(m2·d)−1。 -
潜流型人工湿地种植植物为芦苇,种植密度为16株·m−2。曝气装置安装在进水端,曝气条件为(1.0±0.2) L·min−1。选用甲醇作为外加碳源,共设置3个处理,即不加甲醇对照C/N为0.9∶1(CW0)、添加甲醇调节C/N为2∶1(CWH)、添加甲醇调节C/N为4∶1(CWC)。在每个供试潜流型人工湿地单元的进水端采用点滴输液器将甲醇均匀地添加至湿地进水中。实验周期为90 d,于2019年7月1日添加碳源,预培养30 d后开始采集水样。实验结束后采集基质样品,首先移除采样点表面的种植土、粗砂,继续深挖砾石填料直至出现水面或显著的水浸痕迹,按照S形取样法,采集5个点的样品混匀,取上层粗砂、细砂、细砾石填料各100 g,装入无菌封口袋,按照相同的取样方法采集不同的点位混匀后作为重复。采集的基质样品用液氮急冻,置于放有冰袋的保温箱中,运至实验室,存放至−20 ℃。
-
预运行30 d后采集水质样品,分别于运行39、48、57、66、75、84、93 d采集进水和出水,水样采集时间为上午10:00,在每个湿地单元的进水口和出水口每隔5 min采集1次,共采集3次作为重复。水质测定指标为COD、
NH+4 、NO−3 、TN。COD采用重铬酸钾法(HJ/T 399-2007)测定,TN浓度使用碱性过硫酸钾消解紫外分光光度法(HJ 636-2012)测定,NH+4 浓度采用纳氏试剂比色法(GB/T 5750-2006)测定,NO−3 浓度采用紫外分光光度计法[18]测定。 -
将采集的填料样品放入塑料杯中,加超纯水100 mL,用超纯水涡旋振荡10 min洗脱生物膜,取上清液于10 000 r·min−1离心15 min,收集微生物富集物。称取约0.10 g土壤样品和0.10 g洗脱下来的生物富集物,采用E. Z.N.A.Soil DNA Kit (D5625, Omega, Inc., USA)提取。DNA提取步骤按照试剂盒说明书的操作程序进行。最后DNA溶解于60 μL solution 6溶液中。利用NanoDrop ND-1000(Thermo Scientific,Wilmington,DE)测定DNA浓度、检测DNA质量。
-
利用实时定量PCR检测仪(Bio-Rad, USA)测定功能基因丰度,anammox 16S rRNA、amoA、nxrA、nirK、nirS、nosZ引物序列[19-23]如表1所示。引物序列由金斯瑞生物科技有限公司合成。定量PCR反应液体系为 20 μL,其中包含10 μL SYBR 2 Premix Ex Taq (Takara Shuzo, Shiga, Japan),正向引物和反向引物各0.8 μmol·L−1,0.2 μL牛血清蛋白(BSA, 20 mg·mL−1),2 μL 10倍稀释的DNA(浓度在10~20 ng·μL−1)作为模板,6.2 μL 灭菌水。每个样品设置3个重复。将含有正确目的基因的质粒10倍梯度稀释后作为标准曲线。每次定量均需测定用灭菌水为模板的阴性对照。溶解曲线只出现1个特殊峰。扩增效率在90%~110%才可以应用。
-
采用高通量Illumina MiSeq300 平台测定细菌序列,16S rDNA V3-V4区扩增引物为338F (ACTCCTACGGGAGGCAGCAG),806R(GGACTACHVGGGTWTCTAAT)[21]。用2%琼脂糖凝胶电泳对PCR扩增产物进行检测,检测合格后用DNA凝胶回收试剂盒回收目标片段。扩增产物送至联川生物技术股份有限公司(杭州,中国)进行细菌组分分析。检测合格的序列按照97%的序列相似度聚类成OTU,采用QIIME1.8.0分析细菌群落结构和α多样性,利用RDP(Ribosomal Database Project)对物种进行分类。
-
采用Excel 2013和SPSS 19.0软件对水质数据和功能基因拷贝数进行分析,方差分析和差异显著性比较用单因素(one-way ANOVA)和邓肯(Duncan)法分析,文中所有图采利用Sigmaplot 12.5软件作图。
1.1. 实验地点
1.2. 实验处理与样品采集
1.3. 水质监测
1.4. DNA提取
1.5. 定量PCR
1.6. 高通量测序
1.7. 数据处理
-
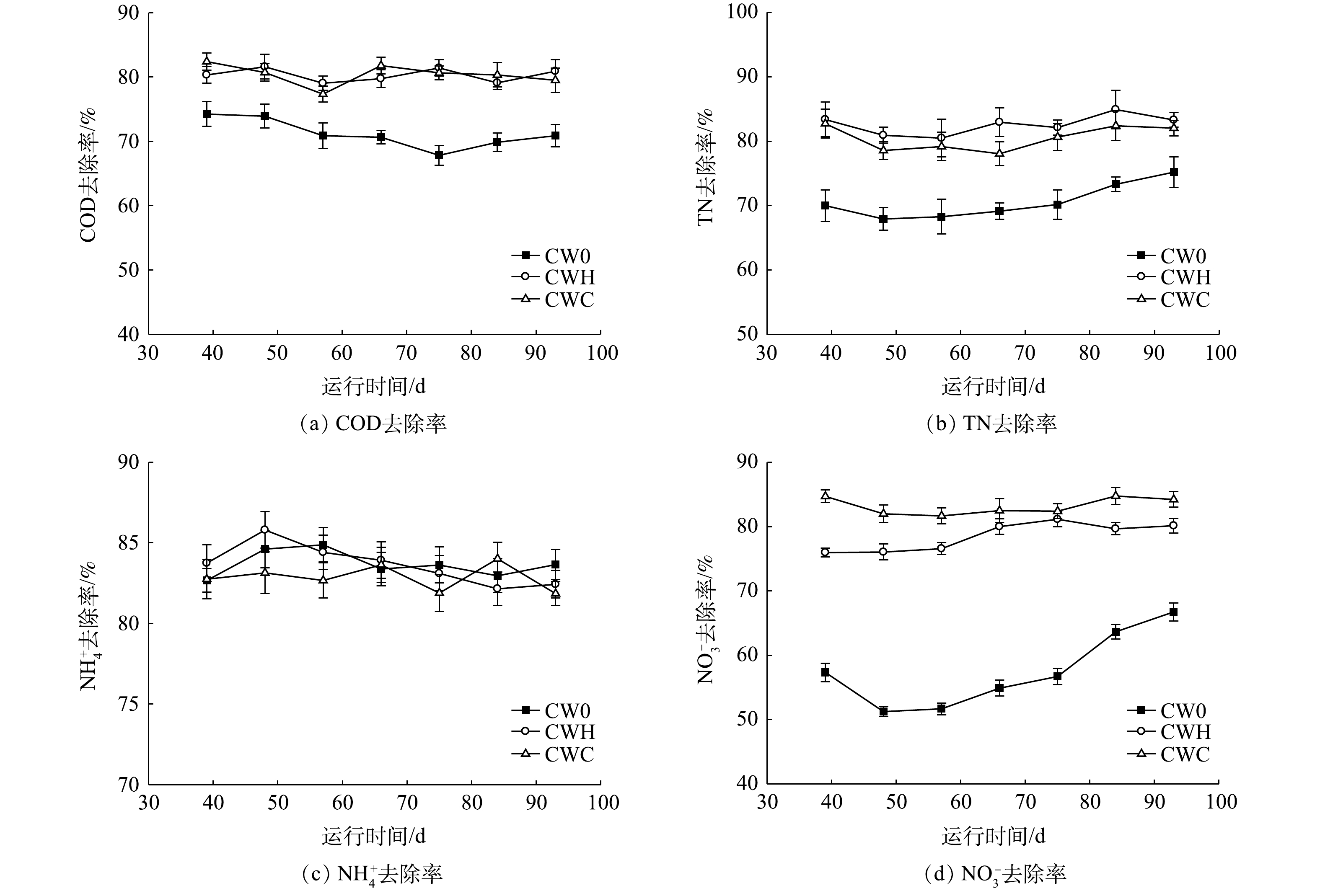
曝气和添加碳源强化措施处理的水平潜流型人工湿地对COD、TN、
NH+4 去除率如图1所示。CW0(C/N=0.9)、CWH(C/N=2)、CWC(C/N=4)的COD平均去除率分别为71.20%、80.32%、80.40%。在曝气条件下添加碳源可增加COD的去除率,CWC、CWH中COD的去除率均显著高于CW0(P<0.05),但CWC与CWH之间无显著差异(P>0.05)。在曝气条件下添加碳源可增加总氮的去除率,各处理的总氮去除率均高于70%,表现为CWH、CWC 的TN平均去除率显著高于CW0(P<0.05),CWH和CWC的TN平均去除率分别为82.59%和80.52%,两者之间无显著差异(P>0.05)。在不同C/N下NH+4 去除率无显著差异(P>0.05),CW0、CWH、CWC的NH+4 平均去除率分别为83.68%、83.65%、82.85%。硝态氮去除率随C/N增加显著增加,CW0、CWH、CWC的NO−3 平均去除率分别为57.48%、78.52%、83.19%。 -
氮转化过程中的主要功能基因丰度如图2所示。CW0、CWC、CWH的anammox 16S rRNA基因丰度分别为5.19、5.67和5.79 log拷贝数·g−1,CWC和CWH处理的anammox 16S rRNA丰度均显著高于CW0(P<0.05),而CWC与CWH 处理间无显著差异(P>0.05)。amoA基因和nxrA基因是硝化过程的标记物,CW0、CWH、CWC的amoA基因和nxrA基因丰度分别为3.83、3.81、4.02 log拷贝数·g−1和3.98、3.68、3.52 log拷贝数·g−1。CW0处理的nxrA基因丰度显著高于CWH和CWC处理(P<0.05),而CWC和CWH处理间无显著差异(P>0.05)。
nirS、nirK、nosZ是反硝化过程的主要功能基因。CW0、CWH、CWC处理的nirS基因丰度分别为3.76、3.79、3.91 log拷贝数·g−1,不同处理间无显著差异(P>0.05)。提高碳氮比显著增加了nirK基因丰度,CW0、CWH、CWC处理的nirK基因丰度分别为3.75、4.71和4.46 log拷贝数·g−1,CWC和CWH处理的nirK基因丰度显著高于CW0处理(P<0.05)。CW0、CWH、CWC处理的nosZ基因丰度分别为2.78、2.65和2.26 log拷贝数·g−1,不同处理的nosZ基因丰度与nirK基因丰度具有相同的趋势,均表现为加碳源处理组显著高于不加碳源的对照组。
-
1)微生物群落丰富度和多样性分析。CWH和CWC处理的物种数均高于CW0,CWC、CWH处理的物种数较CW0增加了13.36%和7.64%(表2)。表征细菌多样性的Shannon-Wiener和Simpson指数、表征群落丰富度的Chao1指数均表现为随C/N增加而增加的趋势,这说明添加碳源增加了物种多样性和丰富度。
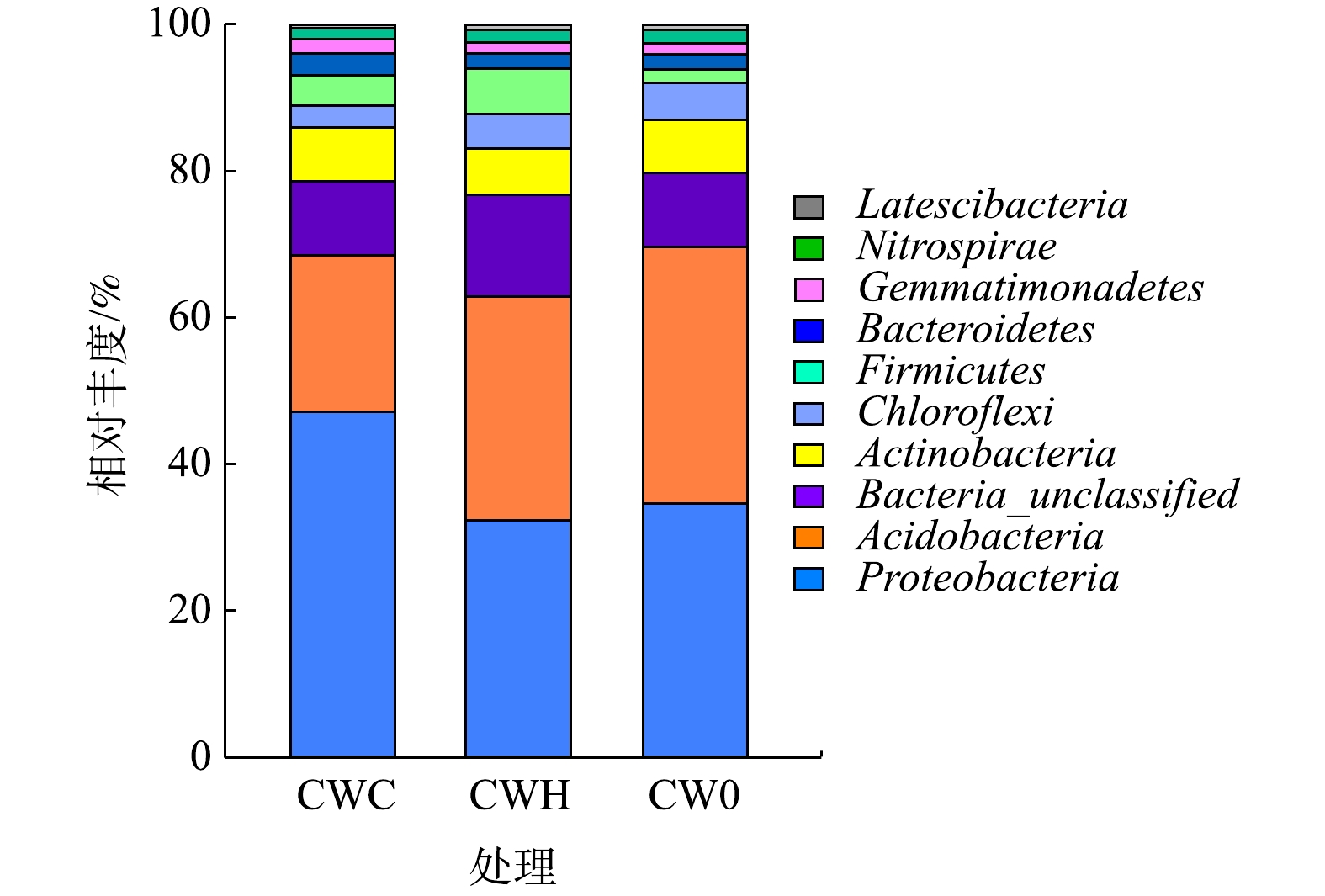
2)细菌在门水平的群落组成。选取丰度排名前10位的物种进行分析(图3)。变形菌门和酸杆菌门相对丰度较高,分别为32.26%~47.11%和21.38%~35.10%。所有处理中均表现为变形菌门丰度最高,CWC处理的变形菌门相对丰度(47.11%)显著高于CWH处理(32.26%)和CW0处理(34.56%)(P<0.05)。而加碳源显著降低了酸杆菌门丰度(P<0.05),CW0处理(35.10%)的酸杆菌门丰度是CWC处理(21.38%)和CWH处理(30.63%)的1.64倍和1.15倍。CWC(4.05%)和CWH(6.14%)的厚壁菌门(Firmicutes)相对丰度均显著高于CW0(1.80%)(P<0.05)。CWC处理的拟杆菌门(Bacteroidetes)相对丰度(2.98%)显著高于CWH处理(2.03%)和CW0处理(2.00%)(P<0.05)。CW0、CWH、CWC的硝化螺旋菌门(Nitrospirae)相对丰度分别为1.87%、1.77%、1.45%,硝化螺旋菌门相对丰度随C/N增加呈逐渐降低趋势。
-
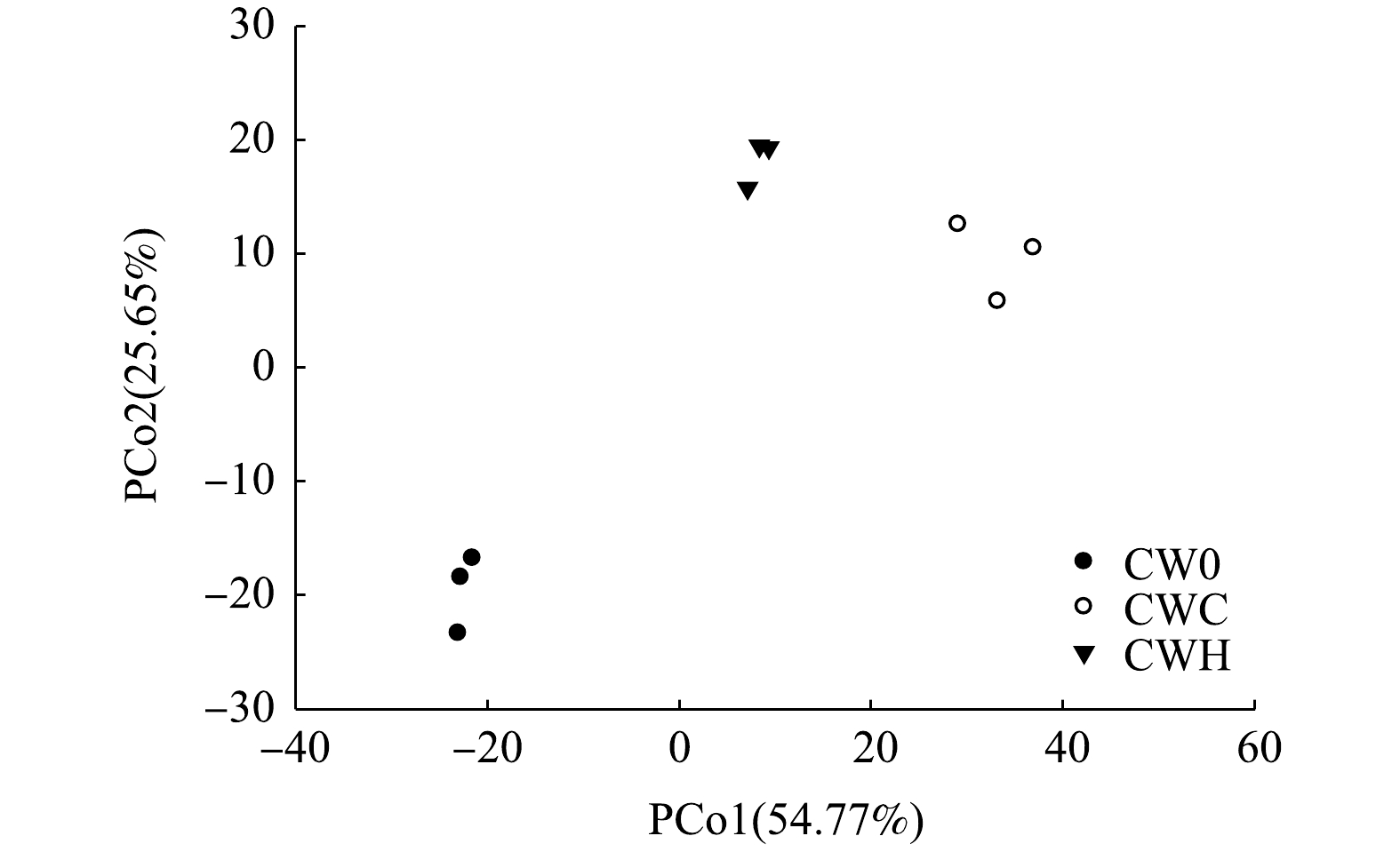
对细菌高通量测序结果(图4)进行PCoA分析发现,加碳源处理(CWC、CWH)与CW0在PCo1和PCo2轴上均距离较远,说明其微生物群落组成结构差异较大,添加碳源对人工湿地基质层微生物群落组成的影响较大。PCo1解释量为54.77%,CWH和与CWC位于PCo1轴的正值端,CW0位于PCo1轴的负值端。PCo2的解释量为25.65%,CWC和CWH位于PCo2轴的正值端,CW0在PCo2轴的负值端。
2.1. 不同处理的湿地中污染物的去除率
2.2. 不同处理下氮转化功能基因丰度
2.3. 不同处理下微生物群落结构分析
2.4. 微生物群落结构差异性分析
-
碳源和可溶性氧是水平潜流型人工湿地中有机质和氮去除的重要影响因子。对曝气条件下低C/N污水的处理结果表明,CW0、CWH、CWC处理的出水中COD无显著差异,这说明在本研究中不同C/N处理均未造成二次污染。
NO−3 去除率随C/N的增加而增加,CW0处理的C/N显著低于CWH和CWC。这说明在低C/N条件下,由于碳源缺乏导致电子供体不足抑制了反硝化过程,随着碳源增加,异养反硝化微生物快速生长繁殖,改善了反硝化速率[10]。CWH和CWC处理的TN去除率显著高于CW0,CWH和CWC处理间无显著差异,说明TN去除率并非随C/N增加而增加,这与CHEN等[24]的研究结果一致。潜流型人工湿地脱氮效率受基质、植物、微生物等多种因素的影响,在人工湿地实际运行过程中,发挥作用的碳源并非只是污水中的碳源。湿地植物根际有机碳释放速率、根系分泌物类型或数量均会影响根系周围微生物丰度和活性[25]。ZHAI等[26]根据湿地植物DOC释放速率推测,每年根系分泌物能够促进人工湿地反硝化脱氮94~267 kg·hm−2。在低碳高硝态氮污水处理中,根系分泌物是潜在的重要碳源。本研究中,供试人工湿地已运行3 a,所种植物芦苇为多年生植物,根系较发达,且该实验在植物生长旺盛期开展,故根系分泌物释放量较高。另外,植物根系残茬腐解亦释放部分碳源,也可以为低C/N污水处理提供碳源[27]。本实验中,在芦苇生长旺盛期,当C/N分别为2和4时,很多指标并未呈现出显著差异。这可能是由于植物根际效应对低C/N污水处理中氮转化功能微生物的影响较大,削弱了外源碳添加导致的处理间的差异。一方面,根系能够为附着的微生物提供较大的表面积,为根际微生物提供适宜的生存条件,因而促进了微生物的生长繁殖[28];另一方面,植物根系分泌物会改变微生物群落结构,提高氨氧化细菌和反硝化细菌丰度[13,29]。本研究仅对低碳氮比条件下人工湿地运行开展了初步的探索,至于植物根际效应与氮转化功能微生物相关性、根系分泌物释放量和植物细根腐解释放碳源等因素与污水处理效果的相关性还有待深入的研究。 -
为了解强化措施对低C/N污水氮转化过程的影响,对硝化、反硝化和厌氧氨氧化过程的主要功能基因进行了分析。本研究中,厌氧氨氧化16S rRNA基因丰度表现为添加碳源处理(C/N为2.0和4.0)显著高于对照处理。厌氧氨氧化过程的发生与底物浓度和厌氧条件有关,添加碳源促进了反硝化过程的发生,底物
NO−2 浓度增加,为厌氧氨氧化过程提供了充足的底物;此外,有机物氧化消耗可溶性氧造成的厌氧条件也促进了厌氧氨氧化过程的发生[10]。本研究中,不同处理的amoA丰度无显著差异,而不加碳源处理的nxrA基因丰度显著高于加碳源处理。这与以前研究结果不一致。大多研究认为,有机物氧化会与氨氧化过程竞争氧气,amoA和nxrA丰度随C/N增加而降低,硝化过程受抑制[11-12]。造成本研究结果与前人研究结果不一致的原因是:前人研究[10-11]中C/N较高(C/N≥6),而本研究中C/N较低(C/N≤4),且进水中可溶性氧含量可以为硝化过程提供部分氧气,并未限制氨氧化过程。nxrA介导的是硝化反应的第二步,加入碳源增加了氧气消耗,而进水中的可溶性氧不足以供应NO−2 氧化为NO−3 过程的发生,因有机物降解与亚硝酸盐氧化过程竞争可溶性氧,NO−2 氧化为NO−3 的过程受抑制[30]。反硝化微生物主要是异养微生物,添加甲醇作为碳源可以促进反硝化脱氮[10]。本研究中,不同处理的nirS基因丰度无显著差异,而CWH、CWC处理的nirK基因丰度显著高于CW0处理,添加碳源对nirS和nirK基因丰度产生不同影响的原因是nirK基因对环境变化较敏感,其丰度受环境因素的影响显著[31]。CWC和CWH处理的nosZ基因丰度均高于CW0处理,nosZ基因通常作为完全反硝化的标志基因,说明添加碳源促进了完全反硝化过程的发生[31]。对潜流型人工湿地基质微生物群落结构的分析表明,添加碳源增加了物种多样性和丰富度。这与LI等[32]的研究结果一致。他们发现,在C/N为2∶1时物种数量最高,在低C/N的条件下,植物根系发达促进了微生物的附着和生长。本研究中,所有处理均表现为变形菌门丰度最高,这与前人的研究[33]结果一致。变形菌门细菌种类繁多,广泛参与碳、氮循环过程。CWH处理的厚壁菌门丰度最高,厚壁菌门能够执行异养反硝化的过程[34],这也是添加碳源能够提高反硝化效率的原因之一。硝化螺旋菌门相对丰度随C/N增加呈逐渐降低趋势,说明添加碳源增加了对氧气的消耗,从而抑制了硝化微生物生长繁殖[12],不同碳氮比对硝化菌组分的影响还有待进一步研究。
3.1. 强化措施对低C/N污水中氮去除率的影响
3.2. 强化措施对氮转化功能微生物丰度和群落结构的影响
-
1)人工湿地在植物生长旺盛期,在曝气条件下调节C/N为2和4均可显著提高TN和
NO−3 的去除率。2)添加碳源改变了氮转化功能基因丰度,在C/N为2和4处理中的nirK、nosZ、厌氧氨氧化细菌16S rRNA基因丰度均显著高于对照。
3)添加碳源可增加物种的丰富度,从而改变细菌群落的结构。
















 DownLoad:
DownLoad: