抗生素能够抑制和杀灭致病菌并且促进畜禽生长,在医疗市场、养殖业占有重要地位。在我国,抗生素生产和使用量都较大,产量达到2.48×105 t·a-1,使用量达到1.62×105 t·a-1;其中48%用于人体医疗,其余专供畜禽养殖[1]。抗生素在畜禽体内无法被完全消化,未吸收的部分以排泄物的形态,经由有机肥施用、污水灌溉等途径,最终进入土壤环境中[2]。土壤环境持续不断地输入大量抗生素,促使抗生素抗性细菌(antibiotic resistant bacteria, ARB)和抗生素抗性基因(antibiotic resistance genes, ARGs)在土壤和动植物之间富集和扩散,威胁农产品质量安全和人体健康[3]。
农田土壤安全问题是事关食品安全和生态平衡的关键问题,已引起广泛关注[4]。土壤是抗生素的重要来源,也是抗性基因的巨大储存库。通过土壤中复杂的微生物活动,ARGs可以在土壤-农作物系统和土壤-土壤动物系统中传播扩散,对农田生态系统产生不可逆的伤害,而且能够通过多种暴露途径(如食物链等)与人体接触,进而富集于人体中,存在较高的安全隐患[5]。目前缺乏携带耐药基因的人畜共生菌在农田生态系统食物链中迁移转化和在食物链各营养级中生命活动的系统认识,因此阐明农田生态系统中耐药菌及其携带的抗性基因在各营养级的定殖规律、扩散现象及转化机制显得尤为重要。
环境中,尤其是与食品安全息息相关的农田中抗生素耐药性引发了不容忽视的生态风险和健康风险。2019年,我国农业农村部发布公告,自2020年起退出除中药外的所有促生长类药物饲料添加剂品种[6]。开展农田生态系统抗生素污染现状调查,同时阐明耐药菌及其携带的抗性基因的研究方法,有利于进一步明确ARGs在农田生态系统中的迁移途径和转化机制,降低抗生素抗性传播风险。本文针对农田生态系统中抗生素抗性基因的研究概况,归纳ARB和ARGs的研究方法,从抗生素抗性基因在农田生态系统各营养级的迁移转化途径进行综述,并对当前研究的不足之处提出进一步研究展望,以期为降低农产品抗性污染提供科学依据。
1 农田生态系统中抗生素抗性基因的来源与传播(Source and spread of antibiotic resistance genes in farmland ecosystem)
地球生态系统中,农作物及其根系周围的土壤环境(土壤、微生物和土壤动物)组成了独特的农田生态系统,是ARGs从环境向人群传播的重要系统[5]。农田土壤环境中普遍存在一些天然抗生素,同时大量耐药微生物和耐药基因常随着施肥等农业活动进入土壤环境,导致耐药基因在畜禽粪便-土壤系统中富集和传播。养猪场附近的农田土壤样本中ARGs的平均富集量是某原始森林的100倍,转座酶的富集量更多达1 000倍[7]。与不施肥相比,配施有机肥显著增加了土壤ARGs的数量和相对丰度,增加幅度分别为110%和91%,表明有机肥施用可能是农田土壤ARGs积累的主要原因[8]。中水回用和大气干湿沉降也是ARGs进入土壤的重要媒介。地表河流和灌溉污水中存在大量ARB和ARGs,经再生水灌溉的公园土壤中富集了147种ARGs,是未经再生水灌溉的土壤的几十甚至上千倍[9]。此外,在医院、养殖场和污水处理厂等环境的空气中都发现了不同程度的ARGs和ARB污染[10]。部分北方城市每克灰尘样品中含有106.40拷贝ARGs,且风会扩大其传播范围[11]。
ARGs不仅可以通过风力、灌溉、施肥等物理方式在土壤传播,还可以由蚯蚓、线虫和螨虫等土壤动物的捕食活动促进ARGs在土壤食物链中的传播[12]。在长期施用有机肥的土壤、线虫和蚯蚓肠道中检测出共享的ARGs,这意味着ARGs具有土壤-线虫-蚯蚓这条短的土壤食物链的营养转移潜力[13]。施肥等农业活动改变了农田中抗性基因及其宿主菌的组成,身处其中的土壤动物也不可避免受到影响。土壤中施用有机肥不仅增加土壤弹尾虫微生物群中的ARGs丰度,捕食弹尾虫的螨虫微生物群中ARGs含量也随之增加[14]。
农田土壤中施加有机肥能够增加植物中ARGs的丰度和多样性。通过分析市场售卖的有机和常规生产的生菜,在生菜的叶际和叶内微生物中检测到了134个ARGs,其中有机生菜的叶际ARGs绝对丰度是传统生菜的8倍[15]。利用从废水池中回收的鸟粪石作为肥料来培植芸薹属植物,在其叶际能够检测到丰富的ARGs,并且在叶际和鸟粪石中存在相同类型的ARGs[16]。农产品尤其可生食的蔬果是抗性基因进入人体最直接的载体,增加了耐药的致病菌传播至人体的风险。新鲜果蔬和即食蔬菜沙拉等都检出链霉素类、磺胺类、四环素类等多种抗性基因[17]。
综上所述,畜禽粪便、中水回用和大气沉降都是ARB和ARGs进入农田土壤的媒介。在此基础上,土壤动物和农田植物可以进一步吸收并富集抗生素,进而通过食物链进入人体,引发健康风险,如图1所示。
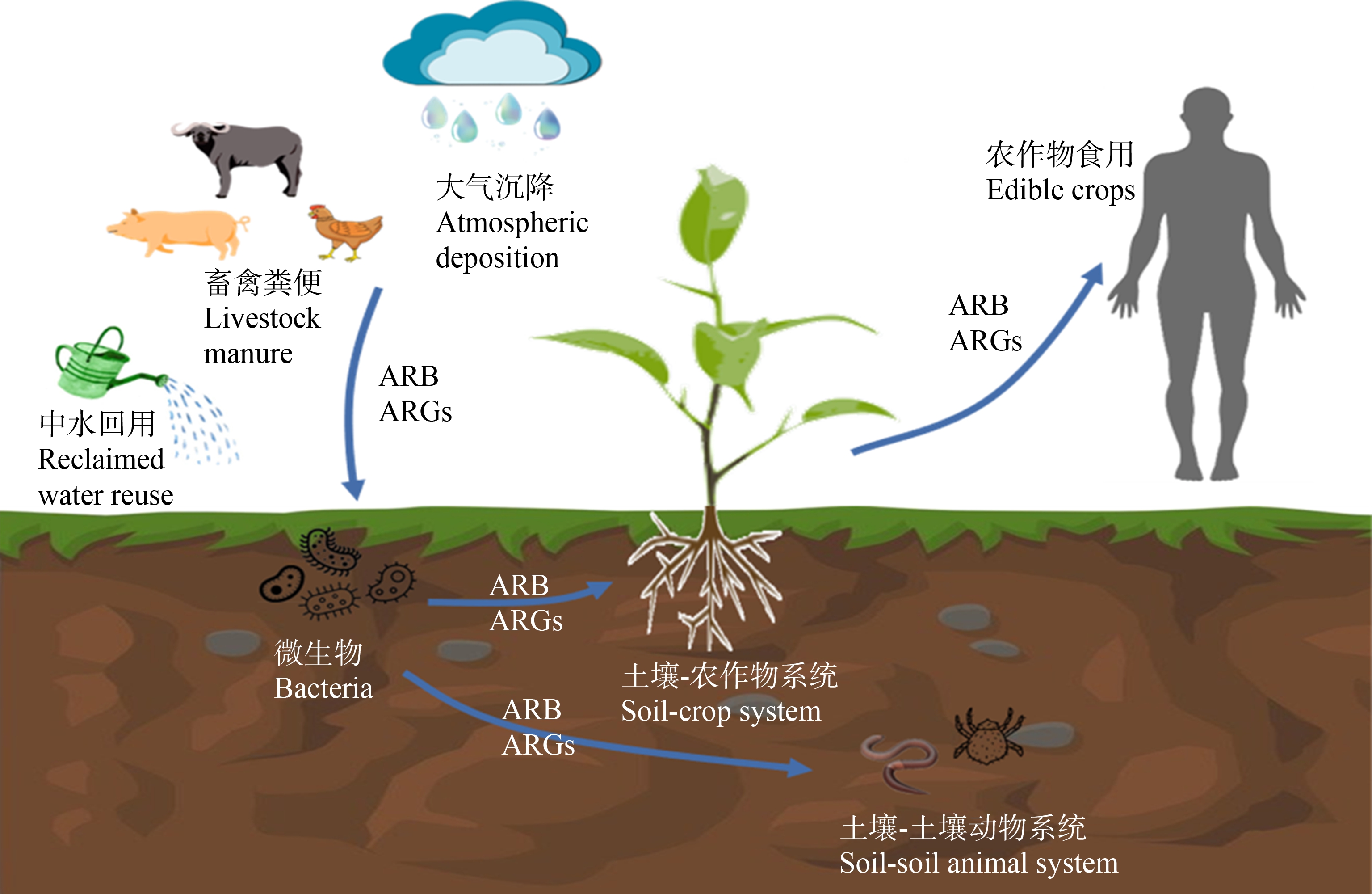
图1 农田生态系统中ARB和ARGs的来源与传播
注:ARB表示抗生素抗性细菌;ARGs表示抗生素抗性基因。
Fig. 1 Source and propagation of ARB and ARGs in farmland ecosystem
Note: ARB stands for antibiotic resistant bacteria; ARGs stands for antibiotic resistance genes.
2 最新国内外文献研究概况(The latest literature research survey at home and abroad)
本文通过中国知网检索关键词“土壤”“抗生素抗性基因”,重点关注近3年来发表的期刊文章,共检索到79篇中文期刊文章,63%为研究性实验,研究的抗生素种类多为四环素和磺胺类,研究方向集中在土壤中ARGs和可遗传移动元件(mobile genetic elements, MGEs)的分布特征,畜禽粪肥和灌溉水等对土壤中ARGs的影响,ARGs与微生物群落的关系等。同理在Web of Science检索“antibiotic resistance genes”“soil”得451篇英文期刊文章,增加“cropland”“farmland”“agricultural”“vegetable”“soil fauna”等关键词组合检索,研究方向主要是ARGs在施肥土壤和蔬菜中的转移,土壤动物对土壤中ARGs的影响,土壤、动物和人之间耐药致病菌的传播等。经文章研读后,筛选并归纳部分满足农田生态系统中ARGs要求的文献如表1所示。
表1 农田生态系统中ARB和ARGs的研究文献归纳
Table 1 Summary of research literature on ARB and ARGs in farmland ecosystem
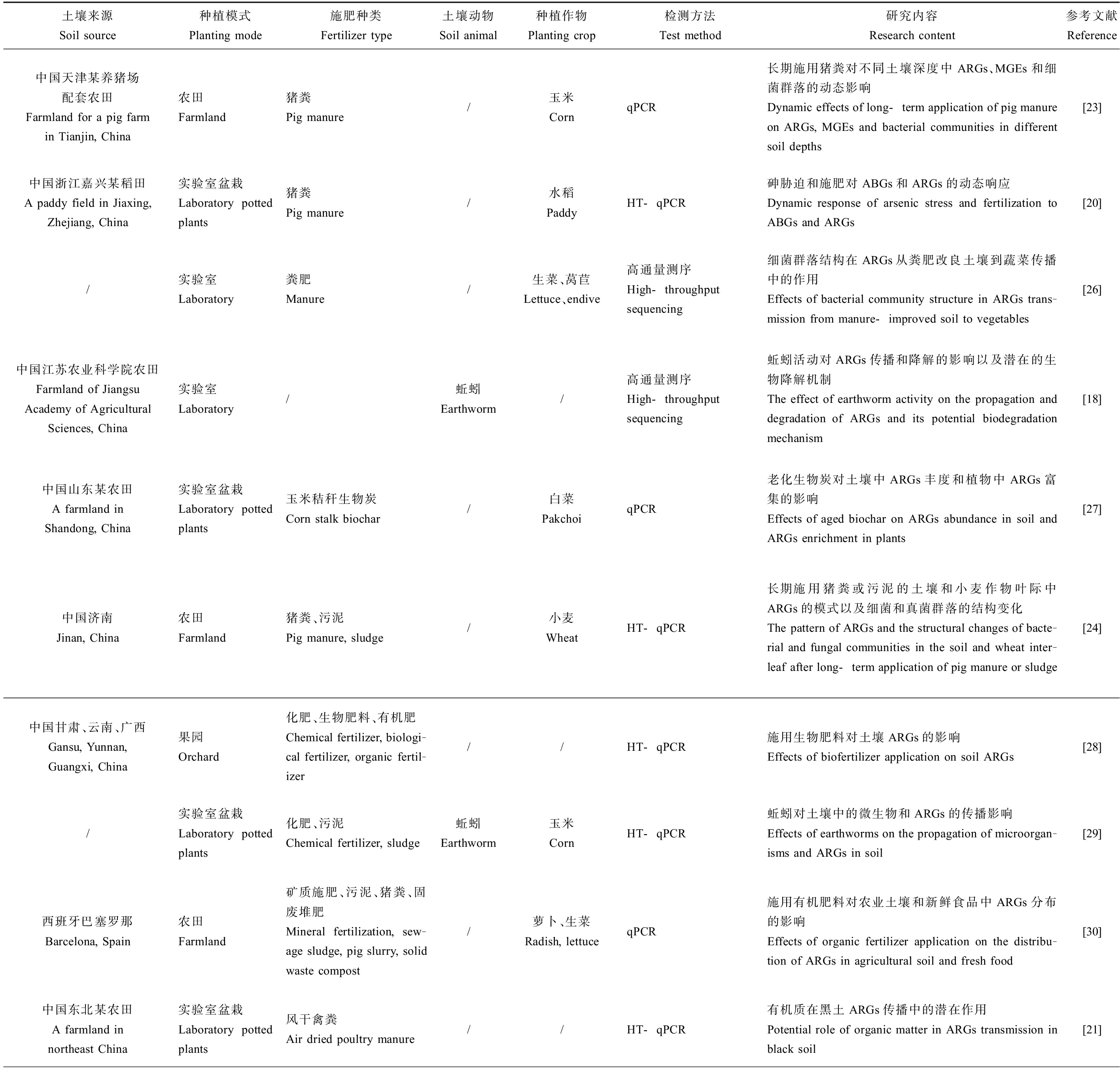
土壤来源Soil source种植模式Planting mode施肥种类Fertilizer type土壤动物Soil animal种植作物Planting crop检测方法Test method研究内容Research content参考文献Reference中国天津某养猪场配套农田Farmland for a pig farm in Tianjin, China农田Farmland猪粪Pig manure/玉米CornqPCR长期施用猪粪对不同土壤深度中ARGs、MGEs和细菌群落的动态影响Dynamic effects of long-term application of pig manure on ARGs, MGEs and bacterial communities in different soil depths[23]中国浙江嘉兴某稻田A paddy field in Jiaxing, Zhejiang, China实验室盆栽Laboratory potted plants猪粪Pig manure/水稻PaddyHT-qPCR砷胁迫和施肥对ABGs和ARGs的动态响应Dynamic response of arsenic stress and fertilization to ABGs and ARGs[20]/实验室Laboratory粪肥Manure/生菜、莴苣Lettuce、endive高通量测序High-throughput sequencing细菌群落结构在ARGs从粪肥改良土壤到蔬菜传播中的作用Effects of bacterial community structure in ARGs trans-mission from manure-improved soil to vegetables[26]中国江苏农业科学院农田Farmland of Jiangsu Academy of Agricultural Sciences, China实验室Laboratory/蚯蚓Earthworm/高通量测序High-throughput sequencing蚯蚓活动对ARGs传播和降解的影响以及潜在的生物降解机制The effect of earthworm activity on the propagation and degradation of ARGs and its potential biodegradation mechanism[18]中国山东某农田A farmland in Shandong, China 实验室盆栽Laboratory potted plants玉米秸秆生物炭Corn stalk biochar/白菜PakchoiqPCR老化生物炭对土壤中ARGs丰度和植物中ARGs富集的影响Effects of aged biochar on ARGs abundance in soil and ARGs enrichment in plants[27]中国济南Jinan, China农田Farmland猪粪、污泥Pig manure, sludge/小麦WheatHT-qPCR长期施用猪粪或污泥的土壤和小麦作物叶际中ARGs的模式以及细菌和真菌群落的结构变化The pattern of ARGs and the structural changes of bacte-rial and fungal communities in the soil and wheat inter-leaf after long-term application of pig manure or sludge[24]中国甘肃、云南、广西Gansu, Yunnan, Guangxi, China果园Orchard化肥、生物肥料、有机肥Chemical fertilizer, biologi-cal fertilizer, organic fertil-izer//HT-qPCR施用生物肥料对土壤ARGs的影响Effects of biofertilizer application on soil ARGs[28]/实验室盆栽Laboratory potted plants化肥、污泥Chemical fertilizer, sludge蚯蚓Earthworm玉米CornHT-qPCR蚯蚓对土壤中的微生物和ARGs的传播影响Effects of earthworms on the propagation of microorgan-isms and ARGs in soil[29]西班牙巴塞罗那Barcelona, Spain农田Farmland矿质施肥、污泥、猪粪、固废堆肥Mineral fertilization, sew-age sludge, pig slurry, solid waste compost/萝卜、生菜Radish, lettuceqPCR施用有机肥料对农业土壤和新鲜食品中ARGs分布的影响Effects of organic fertilizer application on the distribu-tion of ARGs in agricultural soil and fresh food[30]中国东北某农田A farmland in northeast China实验室盆栽Laboratory potted plants风干禽粪Air dried poultry manure//HT-qPCR有机质在黑土ARGs传播中的潜在作用Potential role of organic matter in ARGs transmission in black soil[21]
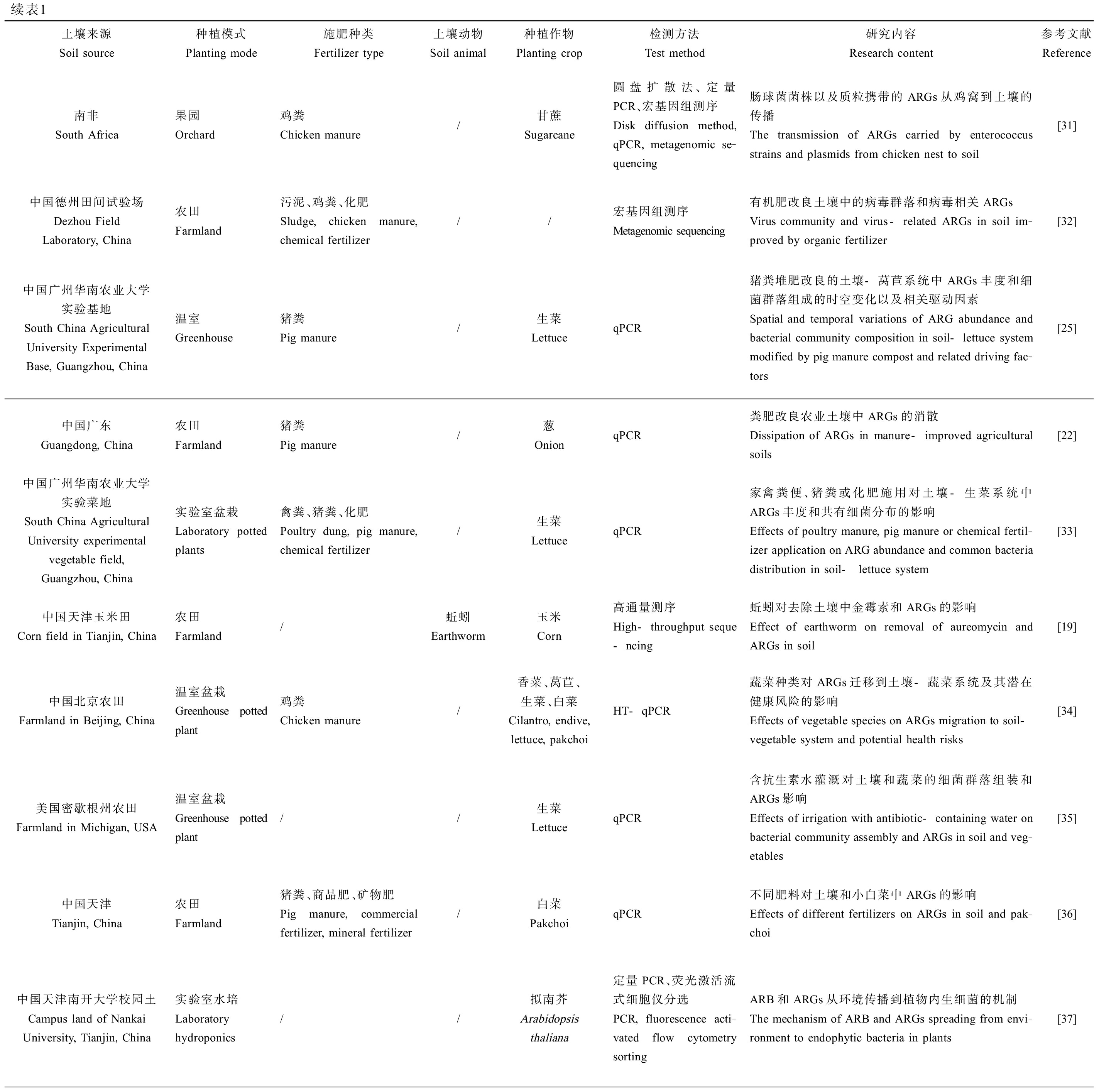
续表1土壤来源Soil source种植模式Planting mode施肥种类Fertilizer type土壤动物Soil animal种植作物Planting crop检测方法Test method研究内容Research content参考文献Reference南非South Africa果园Orchard鸡粪Chicken manure/甘蔗Sugarcane圆盘扩散法、定量PCR、宏基因组测序Disk diffusion method, qPCR, metagenomic se-quencing肠球菌菌株以及质粒携带的ARGs从鸡窝到土壤的传播The transmission of ARGs carried by enterococcus strains and plasmids from chicken nest to soil[31]中国德州田间试验场Dezhou Field Laboratory, China农田Farmland污泥、鸡粪、化肥Sludge, chicken manure, chemical fertilizer//宏基因组测序Metagenomic sequencing有机肥改良土壤中的病毒群落和病毒相关ARGsVirus community and virus-related ARGs in soil im-proved by organic fertilizer[32]中国广州华南农业大学实验基地South China Agricultural University Experimental Base, Guangzhou, China温室Greenhouse猪粪Pig manure/生菜LettuceqPCR猪粪堆肥改良的土壤-莴苣系统中ARGs丰度和细菌群落组成的时空变化以及相关驱动因素Spatial and temporal variations of ARG abundance and bacterial community composition in soil-lettuce system modified by pig manure compost and related driving fac-tors[25]中国广东Guangdong, China农田Farmland猪粪Pig manure/葱OnionqPCR粪肥改良农业土壤中ARGs的消散Dissipation of ARGs in manure-improved agricultural soils[22]中国广州华南农业大学实验菜地South China Agricultural University experimental vegetable field, Guangzhou, China实验室盆栽Laboratory potted plants禽粪、猪粪、化肥Poultry dung, pig manure, chemical fertilizer/生菜LettuceqPCR家禽粪便、猪粪或化肥施用对土壤-生菜系统中ARGs丰度和共有细菌分布的影响Effects of poultry manure, pig manure or chemical fertil-izer application on ARG abundance and common bacteria distribution in soil- lettuce system[33]中国天津玉米田Corn field in Tianjin, China农田Farmland/蚯蚓Earthworm玉米Corn高通量测序High-throughput seque-ncing蚯蚓对去除土壤中金霉素和ARGs的影响Effect of earthworm on removal of aureomycin and ARGs in soil[19]中国北京农田Farmland in Beijing, China温室盆栽Greenhouse potted plant鸡粪Chicken manure/香菜、莴苣、生菜、白菜Cilantro, endive, lettuce, pakchoiHT-qPCR蔬菜种类对ARGs迁移到土壤-蔬菜系统及其潜在健康风险的影响Effects of vegetable species on ARGs migration to soil-vegetable system and potential health risks[34]美国密歇根州农田Farmland in Michigan, USA温室盆栽Greenhouse potted plant//生菜LettuceqPCR含抗生素水灌溉对土壤和蔬菜的细菌群落组装和ARGs影响Effects of irrigation with antibiotic-containing water on bacterial community assembly and ARGs in soil and veg-etables[35]中国天津Tianjin, China农田Farmland猪粪、商品肥、矿物肥Pig manure, commercial fertilizer, mineral fertilizer/白菜PakchoiqPCR不同肥料对土壤和小白菜中ARGs的影响Effects of different fertilizers on ARGs in soil and pak-choi[36]中国天津南开大学校园土Campus land of Nankai University, Tianjin, China实验室水培Laboratory hydroponics//拟南芥Arabidopsis thaliana定量PCR、荧光激活流式细胞仪分选PCR, fluorescence acti-vated flow cytometry sortingARB和ARGs从环境传播到植物内生细菌的机制The mechanism of ARB and ARGs spreading from envi-ronment to endophytic bacteria in plants[37]
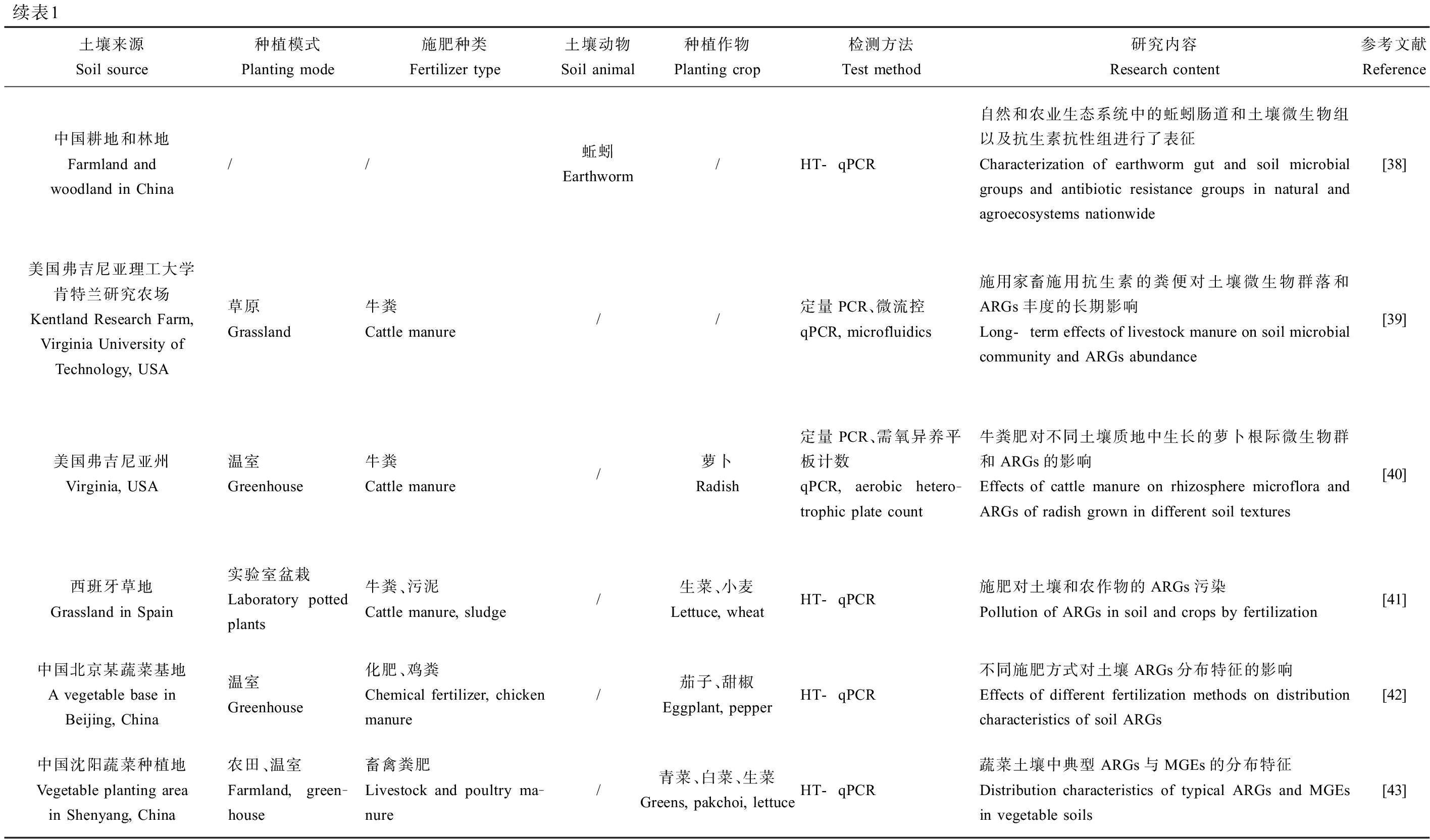
续表1土壤来源Soil source种植模式Planting mode施肥种类Fertilizer type土壤动物Soil animal种植作物Planting crop检测方法Test method研究内容Research content参考文献Reference中国耕地和林地Farmland and woodland in China//蚯蚓Earthworm/HT-qPCR自然和农业生态系统中的蚯蚓肠道和土壤微生物组以及抗生素抗性组进行了表征Characterization of earthworm gut and soil microbial groups and antibiotic resistance groups in natural and agroecosystems nationwide[38]美国弗吉尼亚理工大学肯特兰研究农场Kentland Research Farm, Virginia University of Technology, USA草原Grassland牛粪Cattle manure//定量PCR、微流控qPCR, microfluidics施用家畜施用抗生素的粪便对土壤微生物群落和ARGs丰度的长期影响Long-term effects of livestock manure on soil microbial community and ARGs abundance[39]美国弗吉尼亚州Virginia, USA温室Greenhouse牛粪Cattle manure/萝卜Radish定量PCR、需氧异养平板计数qPCR, aerobic hetero-trophic plate count牛粪肥对不同土壤质地中生长的萝卜根际微生物群和ARGs的影响Effects of cattle manure on rhizosphere microflora and ARGs of radish grown in different soil textures[40]西班牙草地Grassland in Spain实验室盆栽Laboratory potted plants牛粪、污泥Cattle manure, sludge/生菜、小麦Lettuce, wheatHT-qPCR施肥对土壤和农作物的ARGs污染Pollution of ARGs in soil and crops by fertilization[41]中国北京某蔬菜基地A vegetable base in Beijing, China温室Greenhouse化肥、鸡粪Chemical fertilizer, chicken manure/茄子、甜椒Eggplant, pepperHT-qPCR不同施肥方式对土壤ARGs分布特征的影响Effects of different fertilization methods on distribution characteristics of soil ARGs [42]中国沈阳蔬菜种植地Vegetable planting area in Shenyang, China农田、温室Farmland, green-house畜禽粪肥Livestock and poultry ma-nure/青菜、白菜、生菜Greens, pakchoi, lettuceHT-qPCR蔬菜土壤中典型ARGs与MGEs的分布特征Distribution characteristics of typical ARGs and MGEs in vegetable soils[43]
注:ARB表示抗生素抗性细菌,ARGs表示抗生素抗性基因,PCR表示聚合酶链式反应,qPCR表示实时荧光定量聚合酶链式反应,HT-qPCR表示高通量荧光定量聚合酶链式反应。
Note: ARB stands for antibiotic resistant bacteria; ARGs stands for antibiotic resistance genes; PCR stands for polymerase chain reaction; qPCR stands for real-time quantitative polymerase chain reaction; HT-qPCR stands for high-throughput quantitative polymerase chain reaction.
其中,有15.38%的文献探讨了土壤动物对ARGs在土壤中的扩散和降解作用。特征土壤动物蚯蚓加速了四环素类抗生素在土壤中的扩散,但是其对抗生素降解的刺激超过了扩散,而室内实验表明蚯蚓通过刺激潜在的抗生素降解菌和改变土壤微生物群落特性促进了ARGs的降解,其肠道内丰富的谷胱甘肽转移酶可能在ARGs的降解中起到重要作用[18-19]。同时有34.62%研究的是施用畜禽粪便对农田土壤中ARGs和ARB的丰度和多样性的影响。研究表明,粪肥是ARGs和ARB的主要储存库,长期施肥促进了高危ARGs在土壤中的积累和持续存在,同时土壤中重金属、有机质、水分对ARGs和微生物群落产生选择作用[20-22]。在表层土中细菌群落是ARGs变化的主要驱动因素,随着土壤深度的增加,土壤性质对ARGs的影响更为显著[23]。此外,50%探究了ARGs和ARB从粪肥改良的土壤向蔬菜转移。从土壤-蔬菜系统中ARGs的时空分布可以发现,蔬菜内部ARGs的变化主要与其叶内宿主细菌的变化有关。蔬菜过滤掉了土壤中大部分细菌,但富集了叶内层的目标ARGs,且叶际ARGs的相对丰度与MGEs的相对丰度呈正相关[24-25]。而且蔬菜品种还显著影响了根际土壤细菌群落,蔬菜根内生菌、叶内生菌和叶际细菌群落组成差异明显,根内生菌和土壤微生物之间发现了更高丰度的ARGs和共享细菌群落[26]。文献选用的农田大多施用猪粪、牛粪和鸡粪,模式土壤动物多为蚯蚓,研究的蔬菜是以生菜等可生食的叶菜类蔬菜为主。这表明目前国内外不仅研究农田土壤等环境介质中ARB和ARGs的丰度和多样性,而且逐步探索农田生态系统中抗性基因通过各种途径进入食物链的传播机制和潜在风险。
3 农田生态系统中ARGs和ARB定殖的研究方法(Research methods of ARGs and ARB colonization in farmland ecosystem)
3.1 农田生态系统中ARGs的研究方法
农田生态系统常用的ARGs研究方法大体可以概括为微生物培养法、聚合酶链式反应(polymerase chain reaction, PCR)和宏基因组测序三大类。本文总结的国内外文献中,微生物培养法、PCR技术和宏基因组测序的使用情况如图2所示。其中,左侧横坐标图表示3种方法的总使用量,在总文献中分别占15.38%、84.62%和19.23%。而右侧纵坐标图及下方点格图则说明每种研究方法单独或交叉使用情况,69.23%的文献研究单独使用了PCR技术,单独使用微生物培养法和宏基因组测序的研究分别占0%和15.38%,结合2种方法的研究占11.54%,而结合3种方法的仅占3.85%。PCR技术成为现阶段应用最广泛的ARGs检测方法,同时传统的微生物培养法也在发展用以辅助分析耐药菌的表达,而宏基因组测序的出现极大拓展了ARGs的研究思路。

图2 农田生态系统中ARGs研究方法的使用分析图
Fig. 2 Application analysis diagram of ARGs research method in farmland ecosystem
3.1.1 微生物培养法
传统的农田生态系统耐药性研究主要是通过药敏实验,根据微生物耐药表型来评价其抗生素耐药性。常见的微生物培养法有肉汤稀释法、平板稀释法、K-B琼脂扩散法和E-test试验法。目前大多采用K-B琼脂扩增法判断抗性强弱,但是该法不能直接读取细菌的最小抑制浓度(minimal inhibitory concentration, MIC)值[44]。肉汤稀释法和琼脂稀释法可测定MIC值,E-test法将扩散法和稀释法结合,弥补了K-B琼脂法不能测量MIC值的缺点,且操作简便,但是成本高,常用作真菌的药敏试验[45]。有研究结合了K-B琼脂扩散法和肉汤稀释法检测从肉鸡中分离出的禽致病性大肠杆菌分离株对多种抗生素的耐药率,并使用了8种不同的抗菌剂对其进行最低抑菌浓度检测[46]。传统的药敏实验广泛应用于可培养细菌的抗生素抗性检测,但是部分微生物难以进行培养或培养之后无法进行分离,因此难以用于不可培养菌携带的耐药性检测,应用范围和深度有限。
3.1.2 PCR技术
现阶段环境样品中耐药基因检测的主要方法是体外扩增DNA片段的聚合酶链式反应。普通PCR方法无需分离培养就可快速、准确检测到样品中ARGs。同时实时荧光定量PCR(real-time quantitative PCR, qPCR)技术的发展弥补了普通PCR无法定量分析的不足,且因结果稳定,高度自动化和具有实时性,在耐药基因的定量分析中占据主要地位。高通量荧光定量PCR(high-throughput quantitative PCR, HT-qPCR)技术在此基础上大大增加检测样品数量和ARGs种类,同时结合16S rRNA测序研究微生物多样性,显著提升了耐药菌研究的效率和深度,可以更加直观地探究ARGs在环境中分布特征及迁移规律[47]。在不同规模蛋鸡养殖场中采用高通量荧光定量法检测到所有养殖期鸡粪中ARGs亚型132~168种,共存ARGs有110种,而养鸡场周边及施用鸡粪的土壤中共检测到ARGs亚型23~105种,揭示了该养鸡场鸡粪和养殖场周边土壤中抗生素和ARGs的分布特征[48]。但是PCR需要获得目的抗性基因的引物序列,因此环境中未知ARGs的检测受到限制[49]。
3.1.3 宏基因组测序技术
宏基因组测序通过对特定环境样品中提取全部微生物的DNA测全序,与已知的抗生素耐药基因数据库进行序列比对,从而鉴定耐药基因。其核心技术是高通量测序技术,能同时快速检测大量DNA样品,进而研究微生物群落结构及其与环境因子之间的相互作用[50]。宏基因组测序的发展打破了微生物研究的瓶颈,极大地拓展了微生物学的研究思路和研究方法,越来越多地应用于农田土壤生态系统中ARGs的检测和健康风险评估,为研究农田土壤生态系统中ARGs的分布特征和水平转移提供方法支持。利用宏基因组文库进行抗生素耐药性功能筛选也是从环境中获得新耐药基因的一个重要手段[51]。Su等[52]采用宏基因组学方法总共检测到南海140个ARGs亚型和155个毒力因子(virulence factors, VFs),对南海ARGs和VFs的全貌及其丰度和分布进行了研究,而不仅仅局限于几种典型ARGs的检测和丰度。但是由于宏基因组测序的读长较短,容易在同源序列比对时丢失新型ARGs [53-54]。
3.2 农田生态系统中ARB定殖的研究方法
长期使用大量抗生素不仅会致使细菌产生耐药性,同时ARB的存在会加速ARGs在微生物中转移。许多研究表明,MGEs可以将ARGs向已知的人畜共生菌和病原体中转移[55]。因此,有必要探究实际农业活动中是否能通过植物根系内生化和叶际吸附等作用,将环境中携带ARGs的人畜共生菌或致病菌定殖到农产品尤其是可生食蔬果中,造成潜在健康的风险。
3.2.1 内生菌接种方法和影响因素
提高携带耐药基因的内生菌施入土壤后的定殖存活和繁衍迁移能力,有助于对质粒介导ARGs的传播过程进行示踪检测,因此对影响外源微生物定殖因素的研究具有重要作用。不同接种方式会对微生物的定殖量有明显的影响。植物的自然孔口或者外力导致的伤口是微生物定殖到植物体内的关键途径,人工接种就是依靠这些孔口促进微生物进入并定殖到植物组织内部[56]。现在使用较多的植物内生菌接种方法主要包括浸种法、浸根法、涂茎法和叶面喷洒法。鉴于植物根系吸收能力较强且微生物活动丰富,根部的定殖率相对较高,但是同一种接种方法对不同植物的接种效果也会有差异。如采用浇灌法、拌种法和浸根法接种解淀粉芽孢杆菌,发现浇灌法在白菜体内的定殖密度最高,而用浇灌法在烟草体内的定殖却明显小于白菜[57]。
植物可以分泌特异性根际分泌物(包括氨基酸、有机酸等),而微生物能凭借自身的遗传特性(如鞭毛、细胞壁溶解酶等)感知这些分泌物并向根际迁移,最后经过根际环境筛选和微生物内部竞争进行定殖。因此植物的种类和品种、释放的分泌物类别和丰度、微生物的自身遗传特性、土著微生物区系的拮抗竞争作用和土壤的理化性质等因素共同影响了内生菌的定殖能力。在实际的应用中,应该根据研究的农田土壤性质、植物种类和优势菌种类等选择易于定殖和生长的目标微生物和定殖率高的接种方式。
3.2.2 微生物定殖的检测
现在应用于定殖检测的分子生物技术主要有免疫学方法、PCR技术和外源基因标记。免疫学方法采用相应的抗体与目标微生物抗原特异性结合,从而鉴别出目标微生物,是研究微生物定殖的有效方法。基于免疫分析的方法,可以同时检测食源性致病菌大肠杆菌和沙门氏菌,其检出限低达101 CFU·mL-1[58]。PCR技术是用特定的DNA片段检测接种的目标微生物,准确度较高。例如通过对枯草芽胞杆菌全基因组序列比对分析,获得该菌株独有基因序列,设计出针对枯草芽胞杆菌的特异性引物和探针,再利用荧光定量PCR技术和菌落计数法检测枯草芽胞杆菌在叶片上定殖动态[59]。
随着研究需求的提升和生物技术的进步,基因标记法逐渐发展起来,弥补免疫学和PCR技术不能显示微生物活性的不足。基因标记是指将外源DNA插入到受体菌株并稳定表达,与土著微生物产生区别。针对不同微生物种类、不同研究目的可以构建具有生物发光基因、抗生素抗性基因、生化显色基因和铁载体受体基因等不同特征的标记基因系统。在微生物定殖研究中应用最多的则是荧光蛋白标记法。荧光蛋白标记通过导入荧光质粒到受体菌中使受体微生物携带有色荧光,可活体观察,是一种简便灵敏的生物发光标记法[56]。其中,来自海洋多管水母的绿色荧光蛋白(green fluorescent protein, GFP)有着独特的优势,如独具的分子结构、简单的发光机理、稳定的化学性质和不影响生物的正常生命活动等[60]。同时GFP标记的不足之处在于,GFP标记的细胞死亡后不再发出荧光或荧光基因转导不成功会影响检测结果。
3.3 新型检测方法
近些年,随着环境中抗性基因的宿主微生物分离和鉴定的需求增加,推动了许多新型检测方法的发展。基于高通量分离和定向分离2种核心思路,发展了膜扩散培养、微流控培养系统和流式细胞术等方法[61]。用具有延时成像的单细胞微流控技术成功地剖析了质粒介导的水平转移和垂直转移对环境中ARGs传播的贡献[62]。其中流式细胞术不仅能够根据多种表型进行多路分选亚群细胞,而且还能够对混合细胞在单细胞水平上进行多参数定量分析。利用流式细胞术结合荧光标记对荧光假单胞菌进行细胞计数,可以快速简便地检测水中的可同化有机碳[63]。通过纳米双激光系统的流式细胞仪可以分选土壤中提取的红色荧光的供体菌和绿色荧光的转缀合菌[64]。此外,流式细胞术结合单细胞测序技术可以直接大量鉴定和组装环境中未可培养的细菌,从而可以从农田生态系统的ARB中鉴定各种参与特定功能或转移途径的新型基因[65]。
随着第三代测序技术测序读长从短到长,农田环境中ARGs、MGEs及其宿主菌基因序列的检测更加精确[66]。此外,PCR技术和荧光标记结合可以用来研究微生物在农田生态系统中的定殖和迁移。有研究采用PCR定性检测GFP标记的大肠杆菌在小番茄果实、叶片和根系中的定殖和分布[67]。而光电技术的进步使GFP标记与激光共聚焦扫描显微镜相结合可动态地、精确地进行目标微生物繁殖和分布情况的实时跟踪研究,如通过共聚焦显微镜观察GFP标记的大肠杆菌在拟南芥内部的传播[37]。这些新型检测方法从基础研究走到实际应用,将成为研究农田生态系统中抗性基因及其宿主信息极具前景的分析手段。表2对比分析了这些方法的利弊,在实际检测目标耐药菌时,应根据需要将2种或多种方法相结合。
表2 农田生态系统中ARGs和ARB研究方法对比分析
Table 2 Comparative analysis of ARGs and ARB research methods in farmland ecosystem
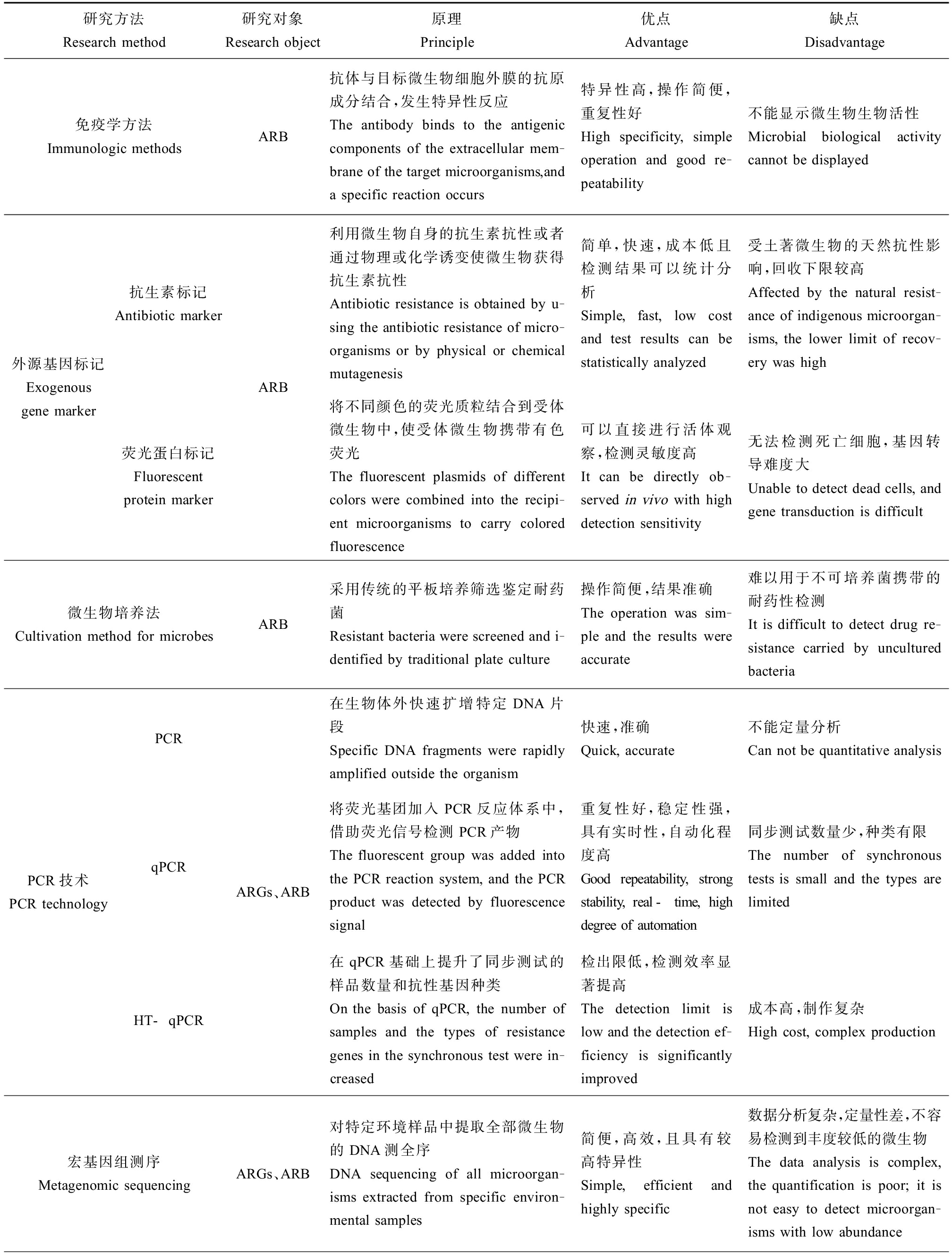
研究方法Research method研究对象Research object原理Principle优点Advantage缺点Disadvantage免疫学方法Immunologic methodsARB抗体与目标微生物细胞外膜的抗原成分结合,发生特异性反应The antibody binds to the antigenic components of the extracellular mem-brane of the target microorganisms,and a specific reaction occurs特异性高,操作简便,重复性好High specificity, simple operation and good re-peatability不能显示微生物生物活性Microbial biological activity cannot be displayed外源基因标记Exogenous gene marker抗生素标记Antibiotic marker荧光蛋白标记Fluorescent protein markerARB利用微生物自身的抗生素抗性或者通过物理或化学诱变使微生物获得抗生素抗性Antibiotic resistance is obtained by u-sing the antibiotic resistance of micro-organisms or by physical or chemical mutagenesis简单,快速,成本低且检测结果可以统计分析Simple, fast, low cost and test results can be statistically analyzed受土著微生物的天然抗性影响,回收下限较高Affected by the natural resist-ance of indigenous microorgan-isms, the lower limit of recov-ery was high将不同颜色的荧光质粒结合到受体微生物中,使受体微生物携带有色荧光The fluorescent plasmids of different colors were combined into the recipi-ent microorganisms to carry colored fluorescence可以直接进行活体观察,检测灵敏度高It can be directly ob-served in vivo with high detection sensitivity无法检测死亡细胞,基因转导难度大Unable to detect dead cells, and gene transduction is difficult微生物培养法Cultivation method for microbesARB采用传统的平板培养筛选鉴定耐药菌Resistant bacteria were screened and i-dentified by traditional plate culture操作简便,结果准确The operation was sim-ple and the results were accurate难以用于不可培养菌携带的耐药性检测It is difficult to detect drug re-sistance carried by uncultured bacteriaPCR技术PCR technologyPCRqPCRHT-qPCRARGs、ARB在生物体外快速扩增特定DNA片段Specific DNA fragments were rapidly amplified outside the organism快速,准确Quick, accurate不能定量分析Can not be quantitative analysis将荧光基团加入PCR反应体系中,借助荧光信号检测PCR产物The fluorescent group was added into the PCR reaction system, and the PCR product was detected by fluorescence signal重复性好,稳定性强,具有实时性,自动化程度高Good repeatability, strong stability, real-time, high degree of automation同步测试数量少,种类有限The number of synchronous tests is small and the types are limited在qPCR基础上提升了同步测试的样品数量和抗性基因种类On the basis of qPCR, the number of samples and the types of resistance genes in the synchronous test were in-creased检出限低,检测效率显著提高The detection limit is low and the detection ef-ficiency is significantly improved成本高,制作复杂High cost, complex production宏基因组测序Metagenomic sequencingARGs、ARB对特定环境样品中提取全部微生物的DNA测全序DNA sequencing of all microorgan-isms extracted from specific environ-mental samples简便,高效,且具有较高特异性Simple, efficient and highly specific数据分析复杂,定量性差,不容易检测到丰度较低的微生物The data analysis is complex, the quantification is poor; it is not easy to detect microorgan-isms with low abundance
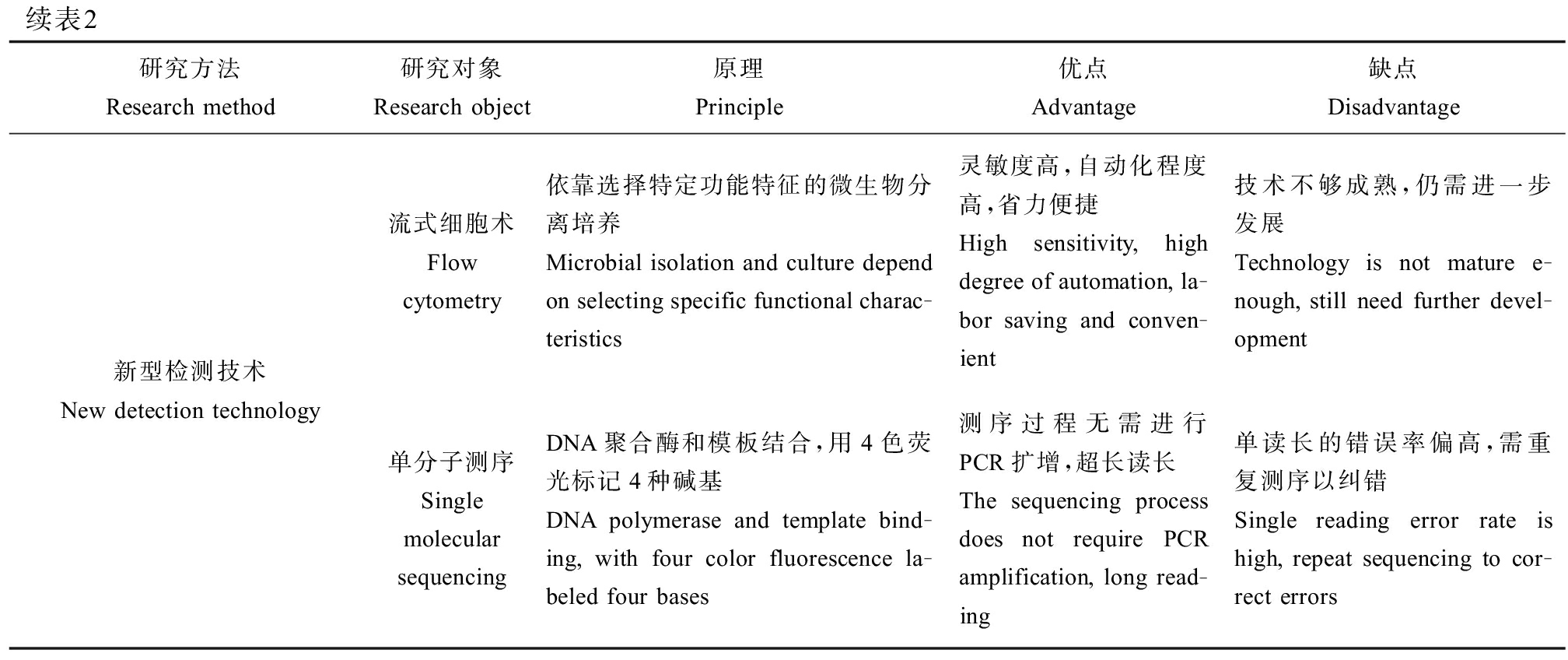
续表2研究方法Research method研究对象Research object原理Principle优点Advantage缺点Disadvantage新型检测技术New detection technology流式细胞术Flow cytometry依靠选择特定功能特征的微生物分离培养Microbial isolation and culture depend on selecting specific functional charac-teristics灵敏度高,自动化程度高,省力便捷High sensitivity, high degree of automation, la-bor saving and conven-ient技术不够成熟,仍需进一步发展Technology is not mature e-nough, still need further devel-opment单分子测序Single molecular sequencingDNA聚合酶和模板结合,用4色荧光标记4种碱基DNA polymerase and template bind-ing, with four color fluorescence la-beled four bases测序过程无需进行PCR扩增,超长读长The sequencing process does not require PCR amplification, long read-ing单读长的错误率偏高,需重复测序以纠错Single reading error rate is high, repeat sequencing to cor-rect errors
4 农田生态系统中ARGs的传播扩散机制(Diffusion mechanism of ARGs in farmland ecosystem)
ARGs在农田生态系统中的传播是一个复杂的过程,从微观分子水平上来说,ARGs的传播方式可以分为垂直基因传递(vetical gene transfer, VGT)和水平基因转移(horizontal gene transfer, HGT)。VGT是指亲代将遗传信息传递给子代,传播范围有限。HGT是指同种或不同种微生物个体间经整合子、转座子、噬菌体及质粒等可移动基因元件介导,以接合、转导和转化等途径传递遗传信息,大大提高了环境中ARGs的迁移扩散效率[68]。因此,HGT被认为是ARGs在农田生态系统中传播扩散的重要原因之一。
土壤ARGs传播中最普遍的转移方式是细菌的接合,细菌间形成的抗性菌毛转移含ARGs的可移动质粒或接合性转座子,因此认为抗性质粒是ARGs水平转移的重要工具。通过GFP标记的耐利福平大肠杆菌捕获来自猪粪细菌的抗生素抗性质粒,并利用PCR检测出81个具有多种抗生素抗性基因的广泛宿主范围质粒[69]。GFP标记质粒RP4从供体菌(恶臭假单胞菌(Pseudomonas putida KT2442))转移到土壤细菌悬浮液,RP4转移发生率最高可以达每104个土壤细菌转移1次,质粒宿主范围广泛,最主要的受体菌为变形菌门[70]。同样有研究探究携带ARGs的RP4质粒及供体菌(恶臭假单胞菌(Pseudomonas putida KT2442))在土壤原位环境中的动态变化,发现质粒的转移效率随培养时间的延长而不断增加,且转移范围不断扩大,受体菌群包括金黄色葡萄球菌等一些潜在人体病原菌[64]。细菌可以直接吸收胞外游离的DNA,从而获得相应遗传性状,这种转化现象是土壤中ARGs转移的另一重要机制。实验室研究表明,包括假单胞菌和不动杆菌在内的近90种细菌存在转化现象[71]。也有研究观察到parC和gyrA基因在肺炎链球菌和草绿色链球菌之间转化[72]。同时抗生素暴露等外界刺激可诱导部分细菌进入“感受态”,从而促使ARGs发生转化[73]。农田土壤存在着大量噬菌体,其感染细菌后可以将遗传物质整合到宿主菌体内并复制,在土壤ARGs转导中起到重要作用。研究发现鸡粪中噬菌体所携带的blaCTX-M和mcr-1抗性基因的含量并不低于质粒[74]。同样的,在蔬菜土壤的噬菌体基因中也检测出β-内酰胺酶类、喹诺酮类等多种ARGs,且传代培养后仍具有再侵染能力,这表明农田土壤中的噬菌体具备携带并传播ARGs的条件[75]。
土壤微生物向植物的迁移也是ARGs在农田生态系统中传播扩散的重要驱动力。将菠菜和甜菜种植在用污水浇灌的农田中,作物体内均检测到了高比例的耐β-内酰胺类内生菌,发现土壤中的ARGs有明显向着植物内生菌系统转移的倾向[76]。最近的拟南芥水培实验进一步证实,供体菌(大肠杆菌(E. coli MG1655))内化到植物组织的同时,供体菌内携带ARGs的RP4质粒转移到植物内生细菌中,更重要的是土壤细菌抑制了大肠杆菌的内化,但显著促进了RP4质粒扩散到植物内生菌中[37]。在粪肥改良的土壤中种植生菜和莴苣,收获时发现土壤中ARGs的丰度明显降低,而蔬菜根系、叶内和叶际微生物中检测到ARGs和Ⅰ类整合子[26]。以上研究表明根际和叶际是ARB传播和HTG发生的热点部位。微生物活动将土壤、植物根系和叶际联系起来,构成ARB传播的内部途径和外部途径。将枯草芽孢杆菌用GFP标记,9 d即可观察到枯草芽孢杆菌成功定殖于白菜根、茎及叶内,且44 d内保持较稳定的定殖量[77]。还有研究发现GFP标记的内生根瘤菌聚集在根际,通过根际的伤口或裂缝进入作物内部,部分根瘤菌还通过维管束系统向上迁移到作物的茎部、叶鞘和叶片中[78]。以上研究证实,土壤中的ARGs通过根系微生物的内生化作用,从根系进入作物组织内部迁移到叶际。此外,土壤ARGs可以通过外部迁移,如堆肥发酵、蒸发等方式,容易被气溶胶化,进而吸附在植株表面[79]。
动物在摄入含ARGs的食物时,其肠道微生物也可能暴露在抗生素和抗性基因污染中。动物肠道内共生着大量微生物,其中部分敏感菌易被其内在或外源ARGs通过HGT等方式诱导成新的耐药菌[80]。如土壤动物在摄食土壤有机质或微生物时,这些ARB和ARGs可能在肠道内定殖并传播到肠道内其他菌群中。施用猪粪不仅让土壤中ARGs的含量增加,还使土壤中ARGs随着线虫、跳虫和捕食性螨的食物链传递[14, 81]。蜜蜂长期暴露在四环素类抗生素污染中,其肠道菌群内也含有四环素抗性基因[82]。也有研究表明接合和转导是哺乳动物肠道微生物水平转移ARGs的主要方式[83]。
以上研究说明,HGT可促进ARGs快速且向更广泛的宿主菌进行迁移,尤其是存在通过食物链转移到人类致病菌中的风险。由图3可知农田生态系统中ARGs的迁移是十分复杂的过程,未来需要更多的研究工作来提供确切的机制阐述。
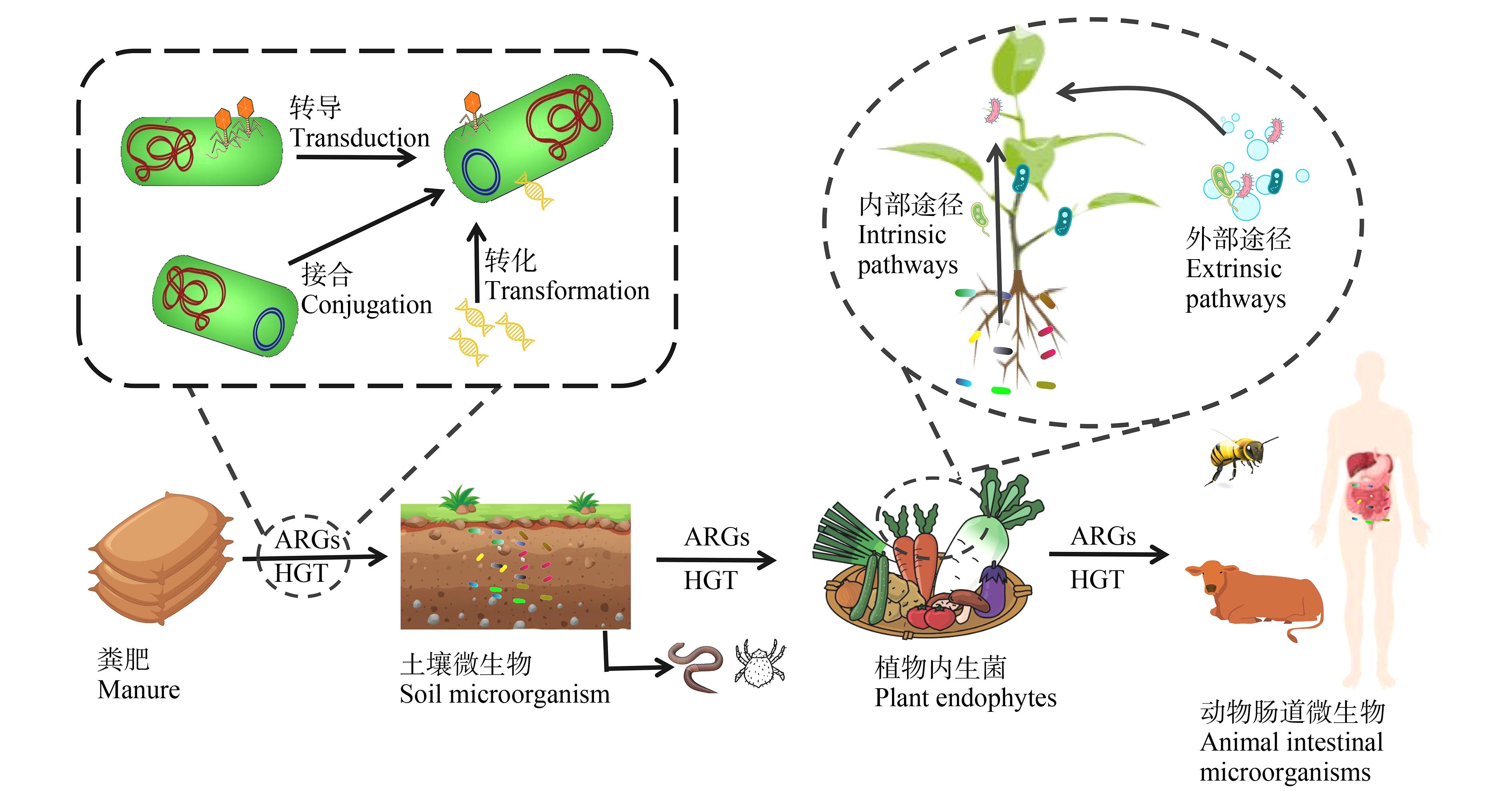
图3 农田生态系统中ARB和ARGs的迁移途径
注:HGT表示水平基因转移。
Fig. 3 Migration pathways of ARB and ARGs in farmland ecosystem
Note: HGT stands for horizontal gene transfer.
5 总结与展望(Summary and prospect)
农田生态系统是人类生存的物质基础,也是受人为活动干扰最多的陆地生态系统。ARB和ARGs不但可以随着施肥等农业活动进入农田土壤,还可以通过入侵植物组织的内部途径和气溶胶等外部途径进一步迁移至农作物。尤其是HGT促进了ARGs在不同物种及环境间转移,甚至转移到了人畜共生菌和潜在病原菌中,存在较高的健康风险。因此研究农田生态系统中ARB和ARGs的分布特征和迁移机制对于降低农产品抗生素抗性污染、保障菜篮子安全具有重要意义。ARB和ARGs的研究方法依旧在不断发展和完善,但研究方向大多集中在调查类的研究,ARGs在农田生态系统中的分布特征、传播途径及转移机制等仍不明晰,目前仍有以下科学问题亟待解决。
(1)现有研究主要关注农田生态系统中抗性基因的丰度和多样性,而可移动部分抗性基因的传播风险仍缺乏足够认识。因此,不同MEGs介导的抗性基因水平转移机制、高频传播和高风险的抗性基因的甄别、关键抗生素抗性基因宿主菌的种类及其ARGs赋存信息、抗性基因转移进入典型人畜共生菌或潜在病原菌的机率等问题仍需进一步研究。
(2)农田生态系统是环境ARGs和耐药病原体向人类传播扩散的重要源之一,但是对ARGs在农田生态系统中尤其是农产品中的转移传播尚不清楚。目前研究已关注到农作物抗性组有多个来源(如土壤、大气环境),但还应关注ARGs不同迁移途径的行为机理及其对农作物抗性组积累的贡献、不同土壤-农作物系统中抗性基因传播的共性和个性,以便针对性地作出ARGs污染控制对策。
(3)食物链是农田生态系统中的ARB和ARGs进入人体的潜在途径。当下对农田生态系统中ARGs的污染风险研究围绕土壤-农作物和土壤-土壤动物展开,对农田生态系统其他营养级的传播途径仍知之甚少。后续可以将食用农作物的土壤动物纳入农田食物链,研究ARGs在土壤-作物-动物系统中的传播扩散,从而为ARGs通过食物链传播至人体的风险评估提供参考。
(4)现代分子生物学技术的发展为环境中抗生素耐药性和抗生素耐药基因的分析提供了更多的可能性,但是不能直观表征ARB和ARGs的扩散传播。因此可以借助GFP荧光标记技术,结合其他研究手段深入探究ARB和ARGs的转移扩散机制。
[1] Zhang Q Q, Ying G G, Pan C G, et al. Comprehensive evaluation of antibiotics emission and fate in the river basins of China: Source analysis, multimedia modeling, and linkage to bacterial resistance [J]. Environmental Science &Technology, 2015, 49(11): 6772-6782
[2] Sun J T, Zeng Q T, Tsang D C W, et al. Antibiotics in the agricultural soils from the Yangtze River Delta, China [J]. Chemosphere, 2017, 189: 301-308
[3] Letten A D, Hall A R, Levine J M. Using ecological coexistence theory to understand antibiotic resistance and microbial competition [J]. Nature Ecology &Evolution, 2021, 5(4): 431-441
[4] 沈仁芳, 颜晓元, 张甘霖, 等. 新时期中国土壤科学发展现状与战略思考[J]. 土壤学报, 2020, 57(5): 1051-1059
Shen R F, Yan X Y, Zhang G L, et al. Status quo of and strategic thinking for the development of soil science in China in the new era [J]. Acta Pedologica Sinica, 2020, 57(5): 1051-1059 (in Chinese)
[5] 朱冬, 陈青林, 丁晶, 等. 土壤生态系统中抗生素抗性基因与星球健康: 进展与展望 [J]. 中国科学: 生命科学, 2019, 49(12): 1652-1663
Zhu D, Chen Q L, Ding J, et al. Antibiotic resistance genes in the soil ecosystem and planetary health: Progress and prospect [J]. Scientia Sinica (Vitae), 2019, 49(12): 1652-1663(in Chinese)
[6] 王娜, 郭欣妍, 单正军, 等. 农田土壤抗生素污染管控建议[J]. 中国工程科学, 2021, 23(1): 167-173
Wang N, Guo X Y, Shan Z J, et al. Suggestions for management and control of antibiotics in farmland soil in China [J]. Strategic Study of CAE, 2021, 23(1): 167-173 (in Chinese)
[7] Zhu Y G, Johnson T A, Su J Q, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms [J]. Proceedings of the National Academy of Sciences of the United States of America, 2013, 110(9): 3435-3440
[8] 冉继伟, 肖琼, 黄敏, 等. 施肥对农田土壤抗生素抗性基因影响的整合分析[J]. 环境科学, 2022, 43(3): 1688-1696
Ran J W, Xiao Q, Huang M, et al. Impacts of fertilization on soil antibiotic resistance genes across croplands: A meta-analysis [J]. Environmental Science, 2022, 43(3): 1688-1696 (in Chinese)
[9] Wang F H, Qiao M, Lv Z E, et al. Impact of reclaimed water irrigation on antibiotic resistance in public parks, Beijing, China [J]. Environmental Pollution, 2014, 184: 247-253
[10] 乔博超, 吴楠, 杨静慧, 等. 环境中抗生素抗性基因的来源、分布及控制对策[J]. 天津农林科技, 2017(6): 16-18
[11] Zhou H, Wang X, Li Z, et al. Occurrence and distribution of urban dust-associated bacterial antibiotic resistance in Northern China [J]. Environmental Science &Technology Letters, 2018, 5(2): 50-55
[12] Popowska M, Rzeczycka M, Miernik A, et al. Influence of soil use on prevalence of tetracycline, streptomycin, and erythromycin resistance and associated resistance genes [J]. Antimicrobial Agents and Chemotherapy, 2012, 56(3): 1434-1443
[13] Zheng F, Bi Q F, Giles M, et al. Fates of antibiotic resistance genes in the gut microbiome from different soil fauna under long-term fertilization [J]. Environmental Science &Technology, 2021, 55(1): 423-432
[14] Zhu D, Xiang Q, Yang X R, et al. Trophic transfer of antibiotic resistance genes in a soil detritus food chain [J]. Environmental Science &Technology, 2019, 53(13): 7770-7781
[15] Zhu B K, Chen Q L, Chen S C, et al. Does organically produced lettuce harbor higher abundance of antibiotic resistance genes than conventionally produced? [J]. Environment International, 2017, 98: 152-159
[16] Chen Q L, An X L, Zhu Y G, et al. Application of struvite alters the antibiotic resistome in soil, rhizosphere, and phyllosphere [J]. Environmental Science &Technology, 2017, 51(14): 8149-8157
[17] Campos J, Mourão J, Pestana N, et al. Microbiological quality of ready-to-eat salads: An underestimated vehicle of bacteria and clinically relevant antibiotic resistance genes [J]. International Journal of Food Microbiology, 2013, 166(3): 464-470
[18] Zheng X X, Chao H Z, Wu Y L, et al. Contrasted effects of Metaphire guillelmi on tetracycline diffusion and dissipation in soil [J]. Journal of Environmental Management, 2022, 310: 114776
[19] Yang S D, Zhao L X, Chang X P, et al. Removal of chlortetracycline and antibiotic resistance genes in soil by earthworms (epigeic Eisenia fetida and endogeic Metaphire guillelmi) [J]. The Science of the Total Environment, 2021, 781: 146679
[20] Cui H L, Zhu D, Ding L J, et al. Co-occurrence of genes for antibiotic resistance and arsenic biotransformation in paddy soils [J]. Journal of Environmental Sciences (China), 2023, 125: 701-711
[21] Li S, Liu J J, Yao Q, et al. Potential role of organic matter in the transmission of antibiotic resistance genes in black soils [J]. Ecotoxicology and Environmental Safety, 2021, 227: 112946
[22] He L Y, He L K, Gao F Z, et al. Dissipation of antibiotic resistance genes in manure-amended agricultural soil [J]. Science of the Total Environment, 2021, 787: 147582
[23] Mu M R, Yang F X, Han B J, et al. Manure application: A trigger for vertical accumulation of antibiotic resistance genes in cropland soils [J]. Ecotoxicology and Environmental Safety, 2022, 237: 113555
[24] Han X M, Hu H W, Li J Y, et al. Long-term application of swine manure and sewage sludge differently impacts antibiotic resistance genes in soil and phyllosphere [J]. Geoderma, 2022, 411: 115698
[25] Wen X, Xu J J, Xiang G F, et al. Multiple driving factors contribute to the variations of typical antibiotic resistance genes in different parts of soil-lettuce system [J]. Ecotoxicology and Environmental Safety, 2021, 225: 112815
[26] Wang F H, Sun R B, Hu H W, et al. The overlap of soil and vegetable microbes drives the transfer of antibiotic resistance genes from manure-amended soil to vegetables [J]. Science of the Total Environment, 2022, 828: 154463
[27] Shi Z M, Zhang P, Liu Y, et al. Accumulation of antibiotic resistance genes in pakchoi (Brassica chinensis L.) grown in chicken manure-fertilized soil amended with fresh and aged biochars [J]. Environmental Science and Pollution Research, 2022, 29(26): 39410-39420
[28] Yang L Y, Zhou S Y, Lin C S, et al. Effects of biofertilizer on soil microbial diversity and antibiotic resistance genes [J]. The Science of the Total Environment, 2022, 820: 153170
[29] Li H, Luo Q P, Pu Q, et al. Earthworms reduce the dissemination potential of antibiotic resistance genes by changing bacterial co-occurrence patterns in soil [J]. Journal of Hazardous Materials, 2022, 426: 128127
[30] Sanz C, Casado M, Navarro-Martin L, et al. Implications of the use of organic fertilizers for antibiotic resistance gene distribution in agricultural soils and fresh food products. A plot-scale study [J]. The Science of the Total Environment, 2022, 815: 151973
[31] Fatoba D O, Amoako D G, Akebe A L K, et al. Genomic analysis of antibiotic-resistant Enterococcus spp. reveals novel enterococci strains and the spread of plasmid-borne Tet(M), Tet(L) and Erm(B) genes from chicken litter to agricultural soil in South Africa [J]. Journal of Environmental Management, 2022, 302(Pt B): 114101
[32] Chen M L, An X L, Liao H, et al. Viral community and virus-associated antibiotic resistance genes in soils amended with organic fertilizers [J]. Environmental Science &Technology, 2021, 55(20): 13881-13890
[33] Huang J L, Mi J D, Yan Q F, et al. Animal manures application increases the abundances of antibiotic resistance genes in soil-lettuce system associated with shared bacterial distributions [J]. The Science of the Total Environment, 2021, 787: 147667
[34] Guo Y J, Qiu T L, Gao M, et al. Diversity and abundance of antibiotic resistance genes in rhizosphere soil and endophytes of leafy vegetables: Focusing on the effect of the vegetable species [J]. Journal of Hazardous Materials, 2021, 415: 125595
[35] Shen Y K, Ryser E T, Li H, et al. Bacterial community assembly and antibiotic resistance genes in the lettuce-soil system upon antibiotic exposure [J]. Science of the Total Environment, 2021, 778: 146255
[36] Xu Y, Li H Y, Shao Z L, et al. Fate of antibiotic resistance genes in farmland soil applied with three different fertilizers during the growth cycle of pakchoi and after harvesting [J]. Journal of Environmental Management, 2021, 289: 112576
[37] Xu H, Chen Z Y, Huang R Y, et al. Antibiotic resistance gene-carrying plasmid spreads into the plant endophytic bacteria using soil bacteria as carriers [J]. Environmental Science &Technology, 2021, 55(15): 10462-10470
[38] Zhu D, Delgado-Baquerizo M, Su J Q, et al. Deciphering potential roles of earthworms in mitigation of antibiotic resistance in the soils from diverse ecosystems [J]. Environmental Science &Technology, 2021, 55(11): 7445-7455
[39] Shawver S, Wepking C, Ishii S, et al. Application of manure from cattle administered antibiotics has sustained multi-year impacts on soil resistome and microbial community structure [J]. Soil Biology and Biochemistry, 2021, 157: 108252
[40] Guron G K P, Chen C Q, Du P, et al. Manure-based amendments influence surface-associated bacteria and markers of antibiotic resistance on radishes grown in soils with different textures [J]. Applied and Environmental Microbiology, 2021, 87(10): e02753-e02720
[41] Jauregi L, Epelde L, Alkorta I, et al. Antibiotic resistance in agricultural soil and crops associated to the application of cow manure-derived amendments from conventional and organic livestock farms [J]. Frontiers in Veterinary Science, 2021, 8: 633858
[42] 左金龙, 孙宇琪, 郭雅杰, 等. 不同施肥处理对蔬菜土壤中抗生素抗性基因多样性与丰度的影响[J]. 环境污染与防治, 2021, 43(5): 553-556, 561
Zuo J L, Sun Y Q, Guo Y J, et al. Effects of different fertilization treatments on the diversity and abundance of antibiotic resistance genes in vegetable soil [J]. Environmental Pollution &Control, 2021, 43(5): 553-556, 561 (in Chinese)
[43] 王百羽, 张珣, 王宝玉, 等. 沈阳蔬菜地土壤中典型抗生素抗性基因与可移动元件分布特征[J]. 生态学杂志, 2021, 40(7): 2113-2119
Wang B Y, Zhang X, Wang B Y, et al. Distribution of typical antibiotic resistance genes and mobile genetic elements in vegetable soils of Shenyang [J]. Chinese Journal of Ecology, 2021, 40(7): 2113-2119 (in Chinese)
[44] McGill K, Kelly L, Madden R H, et al. Comparison of disc diffusion and epsilometer (E-test) testing techniques to determine antimicrobial susceptibiliy of Campylobacter isolates of food and human clinical origin [J]. Journal of Microbiological Methods, 2009, 79(2): 238-241
[45] 周宁, 张建新, 樊明涛, 等. 细菌药物敏感性实验方法研究进展[J]. 食品工业科技, 2012, 33(9): 459-464
Zhou N, Zhang J X, Fan M T, et al. Research progress in antimicrobial susceptibility tests and their applications in bacteria [J]. Science and Technology of Food Industry, 2012, 33(9): 459-464 (in Chinese)
[46] Ibrahim R A, Cryer T L, Lafi S Q, et al. Identification of Escherichia coli from broiler chickens in Jordan, their antimicrobial resistance, gene characterization and the associated risk factors [J]. BMC Veterinary Research, 2019, 15(1): 159
[47] Castiglioni S, Pomati F, Miller K, et al. Novel homologs of the multiple resistance regulator marA in antibiotic-contaminated environments [J]. Water Research, 2008, 42(16): 4271-4280
[48] 沈聪, 张俊华, 刘吉利, 等. 宁夏养鸡场粪污和周边土壤中抗生素及抗生素抗性基因分布特征[J]. 环境科学, 2022, 43(8): 4166-4178
Shen C, Zhang J H, Liu J L, et al. Distribution characteristics of antibiotics and antibiotic resistance genes in manure and surrounding soil of poultry farm in Ningxia [J]. Environmental Science, 2022, 43(8): 4166-4178 (in Chinese)
[49] Allen H K. Antibiotic resistance gene discovery in food-producing animals [J]. Current Opinion in Microbiology, 2014, 19: 25-29
[50] Edwards R A, Rodriguez-Brito B, Wegley L, et al. Using pyrosequencing to shed light on deep mine microbial ecology [J]. BMC Genomics, 2006, 7: 57
[51] Wright G D. The antibiotic resistome: The nexus of chemical and genetic diversity [J]. Nature Reviews Microbiology, 2007, 5(3): 175-186
[52] Su H C, Hu X J, Xu W J, et al. Diversity, abundances and distribution of antibiotic resistance genes and virulence factors in the South China Sea revealed by metagenomic sequencing [J]. The Science of the Total Environment, 2022, 814: 152803
[53] Schmieder R, Edwards R. Insights into antibiotic resistance through metagenomic approaches [J]. Future Microbiology, 2012, 7(1): 73-89
[54] McArthur A G, Waglechner N, Nizam F, et al. The comprehensive antibiotic resistance database [J]. Antimicrobial Agents and Chemotherapy, 2013, 57(7): 3348-3357
[55] 黄丹, 叶茂, 朱国繁, 等. 抗生素/抗性细菌/抗性基因在土壤-植物系统中迁移转化及阻控消减的研究进展[J]. 土壤, 2020, 52(5): 891-900
Huang D, Ye M, Zhu G F, et al. Migration and risk control of antibiotic and antibiotic resistance bacteria/genes in soil-plant system: A review [J]. Soils, 2020, 52(5): 891-900 (in Chinese)
[56] Zhang W C, Li J, Zhi Y W, et al. Research progress on remediation of pollutants in soil using plant-endophyte associations [J]. Journal of Agriculture Resources and Environment, 2021, 38(3): 355
[57] 王志远, 吴兴兴, 吴毅歆, 等. 解淀粉芽孢杆菌B9601-Y2抗性基因标记及其在作物根部的定殖能力[J]. 华中农业大学学报, 2012, 31(3): 313-319
Wang Z Y, Wu X X, Wu Y X, et al. Resistance genes labeling and colonization ability of a biocontrol agent B9601-Y2 of Bacillus amyloliquefaciens in crop rhizospheres [J]. Journal of Huazhong Agricultural University, 2012, 31(3): 313-319 (in Chinese)
[58] Xue L, Huang F C, Hao L, et al. A sensitive immunoassay for simultaneous detection of foodborne pathogens using MnO2 nanoflowers-assisted loading and release of quantum dots [J]. Food Chemistry, 2020, 322: 126719
[59] 刘晓萌, 苏振贺, 宣立峰, 等. 枯草芽胞杆菌HMB19198在番茄叶片上定殖能力的分子检测[J]. 中国生物防治学报, 2022, 38(2): 487-494
Liu X M, Su Z H, Xuan L F, et al. Quantitative detection of Bacillus subtilis HMB19198 on tomato leaf by real-time PCR [J]. Chinese Journal of Biological Control, 2022, 38(2): 487-494 (in Chinese)
[60] 张磊. 草酸青霉菌P8(Penicillium oxalicum)的GFP和潮霉素抗性基因标记及其作物根际定殖的研究[D]. 南京: 南京农业大学, 2005: 20-23
Zhang L. Labelling the phosphate-solubilizing strain P8 of Penicillium oxalicum with GFP and hygromycin resistance genes and its colonizarion in rhizosphere of crops [D]. Nanjing: Nanjing Agricultural University, 2005: 20-23 (in Chinese)
[61] Lewis W H, Tahon G, Geesink P, et al. Innovations to culturing the uncultured microbial majority [J]. Nature Reviews Microbiology, 2021, 19(4): 225-240
[62] Li B, Qiu Y, Song Y Q, et al. Dissecting horizontal and vertical gene transfer of antibiotic resistance plasmid in bacterial community using microfluidics [J]. Environment International, 2019, 131: 105007
[63] Tang P, Wu J, Liu H, et al. Assimilable organic carbon (AOC) determination using GFP-tagged Pseudomonas fluorescens P-17 in water by flow cytometry [J]. PLoS One, 2018, 13(6): e0199193
[64] Fan X T, Li H, Chen Q L, et al. Fate of antibiotic resistant Pseudomonas putida and broad host range plasmid in natural soil microcosms [J]. Frontiers in Microbiology, 2019, 10: 194
[65] Lasken R S. Genomic sequencing of uncultured microorganisms from single cells [J]. Nature Reviews Microbiology, 2012, 10(9): 631-640
[66] Escalona M, Rocha S, Posada D. A comparison of tools for the simulation of genomic next-generation sequencing data [J]. Nature Reviews Genetics, 2016, 17(8): 459-469
[67] 钟薇馨, 石珂弋, 陈意群, 等. 小番茄内生菌耐药基因检测及种植模型中GFP标记菌转移研究[J]. 华南农业大学学报, 2018, 39(4): 55-60
Zhong W X, Shi K Y, Chen Y Q, et al. Detection of antibiotic resistant genes in cherry tomato entophytic bacteria and transfer of GFP marked bacteria in plantation model [J]. Journal of South China Agricultural University, 2018, 39(4): 55-60 (in Chinese)
[68] Dodd M C. Potential impacts of disinfection processes on elimination and deactivation of antibiotic resistance genes during water and wastewater treatment [J]. Journal of Environmental Monitoring: JEM, 2012, 14(7): 1754-1771
[69] Binh C T, Heuer H, Kaupenjohann M, et al. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids [J]. FEMS Microbiology Ecology, 2008, 66(1): 25-37
[70] 邓贝奇. 生菜中抗生素抗性基因污染溯源初探[D]. 杭州: 浙江大学, 2021: 20-22
Deng B Q. Preliminary study on the source tracking of antibiotic resistance genes contamination in lettuce [D]. Hangzhou: Zhejiang University, 2021: 20-22 (in Chinese)
[71] de Vries J, Wackernagel W. Microbial horizontal gene transfer and the DNA release from transgenic crop plants [J]. Plant and Soil, 2005, 266(1): 91-104
[72] Balsalobre L, Ferrándiz M J, Li ares J, et al. Viridans group streptococci are donors in horizontal transfer of topoisomerase Ⅳ genes to Streptococcus pneumoniae [J]. Antimicrobial Agents and Chemotherapy, 2003, 47(7): 2072-2081
ares J, et al. Viridans group streptococci are donors in horizontal transfer of topoisomerase Ⅳ genes to Streptococcus pneumoniae [J]. Antimicrobial Agents and Chemotherapy, 2003, 47(7): 2072-2081
[73] Seitz P, Blokesch M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria [J]. FEMS Microbiology Reviews, 2013, 37(3): 336-363
[74] Ross J, Topp E. Abundance of antibiotic resistance genes in bacteriophage following soil fertilization with dairy manure or municipal biosolids, and evidence for potential transduction [J]. Applied and Environmental Microbiology, 2015, 81(22): 7905-7913
[75] Larra aga O, Brown-Jaque M, Quirós P, et al. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil [J]. Environment International, 2018, 115: 133-141
aga O, Brown-Jaque M, Quirós P, et al. Phage particles harboring antibiotic resistance genes in fresh-cut vegetables and agricultural soil [J]. Environment International, 2018, 115: 133-141
[76] Onalenna O, Rahube T O. Assessing bacterial diversity and antibiotic resistance dynamics in wastewater effluent-irrigated soil and vegetables in a microcosm setting [J]. Heliyon, 2022, 8(3): e09089
[77] 杜芳, 何鹏飞, 吴毅歆, 等. GFP标记内生枯草芽孢杆菌Y10及其在白菜体内的定殖[J]. 生态学杂志, 2015, 34(7): 2064-2070
Du F, He P F, Wu Y X, et al. Colonization of GFP-tagged endophytic Bacillus subtilis Y10 in Chinese cabbage [J]. Chinese Journal of Ecology, 2015, 34(7): 2064-2070 (in Chinese)
[78] Chi F, Shen S H, Cheng H P, et al. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology [J]. Applied and Environmental Microbiology, 2005, 71(11): 7271-7278
[79] Gao M, Jia R Z, Qiu T L, et al. Size-related bacterial diversity and tetracycline resistance gene abundance in the air of concentrated poultry feeding operations [J]. Environmental Pollution, 2017, 220(Pt B): 1342-1348
[80] van Reenen C A, Dicks L M T. Horizontal gene transfer amongst probiotic lactic acid bacteria and other intestinal microbiota: What are the possibilities? A review [J]. Archives of Microbiology, 2011, 193(3): 157-168
[81] Zheng F, Zhu D, Giles M, et al. Mineral and organic fertilization alters the microbiome of a soil nematode Dorylaimus stagnalis and its resistome [J]. The Science of the Total Environment, 2019, 680: 70-78
[82] Tian B Y, Fadhil N H, Powell J E, et al. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees [J]. mBio, 2012, 3(6): e00377-e00312
[83] van Schaik W. The human gut resistome [J]. Philosophical Transactions of the Royal Society of London Series B, Biological Sciences, 2015, 370(1670): 20140087