-
硝化反应是自然界氮循环的关键环节,在氮生物地球化学循环中发挥着重要作用。硝化作用是氨氮(NH4+)首先被氨氧化细菌(AOB)或古菌(AOA)氧化为亚硝态氮(NO2−),进而被亚硝酸盐氧化菌(NOB)氧化为硝态氮(NO3−)的生化反应过程。在过去100多年中,众多学者研究了氨氧化反应过程及AOB菌群的生理特性,而亚硝酸盐氧化及NOB菌群的生理特性并未引起广泛重视。这主要是由于,在传统生物硝化过程中,氨氧化反应是硝化过程的限速步骤,相对于氨氧化反应,亚硝酸盐氧化反应较容易发生[1]。但短程生物脱氮技术的发现,通过不匹配AOB和NOB两类菌属的生理特性差异,选择性抑制NOB菌属的活性,可作为短程生物脱氮的有效途径,因此,众多学者开始关注NOB的菌群特性研究。
NOB属于化能自养型细菌,为专性好氧菌,可利用无机碳化合物,如CO2、CO32−、HCO3−等作为碳源,利用NO2−作为氮源,在NO2−氧化过程中以O2作为最终电子受体,以获得生长所需要的能量。NOB氧化NO2−产生的能量仅有2%~11%用于细胞增殖,因此,NOB能量利用率不高,生长较缓慢,导致其在自然生态系统中的比例较低(4%~5%)[2]。目前,已鉴定出来的NOB菌属主要包括硝化杆菌属(Nitrobacter)、硝化螺菌属(Nitrospira)、硝化球菌属(Nitrococuus)、硝化刺菌属(Nitrospina)、Nitrotoga属、Candidatus Nitromaritima属和Nitrolancea属7类菌属[3]。其中,硝化杆菌属(Nitrobacter)和硝化螺菌属(Nitrospira)是NOB的最典型代表菌属,广泛分布于土壤、淡水、海水及城市污水处理厂活性污泥中。
近10年来,高通量测序技术被广泛用于自然生态系统中微生物群落特性的研究中,能够系统深入地解析、刻画微生物组,通过核酸序列的同源性和差异反映群落的多样性、组成谱和功能谱,并对各种微生物组进行大规模并行比较、关联分析,从而有助于深入探讨它们对于整个生态系统的重要作用,更全面地研究“微生物组-环境/宿主”之间的相互作用机制。
本研究针对富集NOB菌属活性污泥系统,通过SEM技术探究了NOB菌属的形态特征,进而采用宏基因组学技术研究了生化系统内活性污泥的微生物组成及优势物种关联网络关系,探究了NOB菌属的生理生化、代谢活性和行为功能等多方面特性,为自然生态系统氮循环、短程生物脱氮技术提供微生物学理论支持。
-
实验采用人工模拟废水,其进水成分为:10~700 mg·L−1 NaNO2(通过投加不同的NaNO2溶液控制初始的NO2−浓度)、0.4 gNaHCO3、1 gKH2PO4、1.31 gK2HPO4以及0.9 mL微量元素。每升微量元素中:1.25 gEDTA、0.55 gZnSO4·7 H2O,、0.4 gCoCl2·6 H2O、1.275 gMnCl2·4 H2O、0.4 gCuSO4·4 H2O、0.05 gNa2MoO4·2 H2O、1.375 gCaCl2·2 H2O、1.25 gFeCl3·6 H2O、44.4 gMgSO4·7 H2O。接种污泥来自兰州西固兰炼污水处理厂氧化沟好氧段,该污泥具有良好脱氮除磷性能,混合液悬浮固体浓度为3 300 mg·L−1。
实验采用SBR反应器,其有效容积为7 L,控制反应器内混合液反应条件为,溶解氧4~4.5 mg·L−1、温度25±1 °C、pH 7.5±0.5。此外,采用WTW-Multi3420分析仪实时监测硝化反应终点。SBR典型运行周期为:瞬时进水(1 min)、曝气(时间随周期变化)、静置沉淀(60 min)和排水(5 min)。实验共运行140 d(420个周期),进水亚硝酸盐氮质量浓度从10 mg·L−1起步并以50 mg·L−1的梯度逐步提高至700 mg·L−1。
-
NO- 2-N、NO- 3-N、及悬浮固体浓度均采用国家标准方法测定[4-5];pH和溶解氧通过WTW-Multi3420进行实时监测。
-
污泥样品细菌基因组 DNA 采用DNeasyPowerSoil Kit试剂盒提取,并通过0.8%琼脂糖凝胶电泳进行DNA质量检测,同时采用紫外分光光度计对DNA的浓度和纯度进行量化。以细菌16S rRNA的V4-V5区为靶点,进行PCR扩增。扩增引物序列:F为515F(GTGCCAGCMGCCGCGGTAA),R为907R(CCGTCAATTCMTTTRAGTTT)。PCR扩增反应条件为:95 °C预加热2 min;随后进行25周期的扩增反应(95 °C变性30 s、55 °C退火30 s、72 °C延伸40 s);终止后在72 °C下保温10 min,反应液于−80 ℃保存。委托上海派森诺生物科技有限公司进行Illumina MiSeq高通量测序。
-
宏基因组物种注释和丰度分析:1)对高通量测序得到的原始序列进行质量筛查,获取高质量数据集(Clean data);2)对高质量序列进行序列拼接组装,构建宏基因组Contigs和 Scaffolds序列集,并将序列与 NCBI-NT 分析库中的真菌、细菌、古菌和病毒序列进行BLASTN比对(设定E<0.001);3)通过MEGAN[6]利用“最低共同祖先”算法[7]为目标序列的物种分类注释信息;4)结合Saffolds/Scaftigs序列在各组中的丰度数据,获得样本在系统分类等级上的相对丰度表,并进行基因预测,获得非冗余蛋白序列集;5)对蛋白序进行功能注释,获得功能类群丰度谱、代谢通路富集分析等。
-
絮状污泥电镜样品制备方法:1)取污泥混合液于1.5 mL的离心管中,以4 000 r·min−1离心5 min,将上清液去除,加入2.5%戊二醛固定,并置于4 °C冰箱中1.5 h;2)用0.1 mol·L−1的磷酸缓冲液冲洗3次,每次10 min;3)分别用质量分数50%,70%,80%,90%的乙醇进行脱水,每次10~15 min,再以100%的乙醇脱水3次,每次10~15 min;4)100%乙醇∶乙酸异戊酯= 1∶1、纯乙酸异戊酯各置换,每次15 min;5)用HCP22 (H ITACH I)型临界点干燥仪对样品进行干燥;6)在样品表面镀1 500 nm金属膜(金或铂膜)完成预处理。采用扫描电镜(JSM-5 600LV,日本)进行观察并进行能谱分析。
-
为了探究NOB富集系统内微生物主要形态特征,SBR生化系统稳定运行至第120天时,取活性污泥样品采用SEM对其表面形态进行表征(图1)。结果表明,微生物以杆状菌和椭球状菌为主[(0.5~0.8)×(1.0~2.0)],且活性污泥表面形状规则,界限清晰。此外,大量的菌体大都以单体形式存在,但又相互交织在一体,形成了网状的微生物系统。基于相关文献报道的NOB菌属微生物的形态特征,Nitrobacter呈短杆状,Nitrospira呈螺旋状,Nitrococuus呈球状,Nitrospina呈长杆状[8]。
-
基于活性污泥样品微生物在门分类水平群落分布(图2(a)),可以看出,微生物群落在门分类水平上多样性丰富,共检测出19个门。其中相对丰度较高(>1.0%)的门主要包括:变形菌门(Proteobacteria)(82.2%)、放线菌门(Actinobacteria)(4.5%)、拟杆菌门(Bacteroidetes)(4.5%)、厚壁菌门(Firmicutes)(2.1%)、芽单胞菌门(Gemmationadetes)(1.6%)和疣微菌门(Verrucomicrobia)(1.6%),这6类菌门的相对丰度总和约占总生物量的96.5%。其它相对丰度较低(<1.0%)的菌门包括:异常球菌-栖热菌门(Deinococcus-Thermus)(0.9%)、浮霉菌门(Planctomycetes)(0.8%)、绿弯菌门(Chloroflexi)(0.5%)等。NOB的代表性菌属中的Nitrobacter和Nitrospira分别属于Proteobacteria和硝化螺旋菌门(Nitrospirae)。Proteobacteria是自然生态系统细菌界中最大的门,也是活性污泥生化系统中最主要的功能菌群,其相对丰度约为27.5%~65.0%[9-10]。本研究中,Proteobacteria相对丰度(82.2%)高于已有的文献报道值。然而,杨浩等[11]在典型集雨人饮地区窖水微生物群落多样性及差异解析中发现,窖水水样中微生物在门水平上的群落结构中最丰富的是Bacteroidetes(39.1%),其次为Proteobacteria(29.2%)。宁高阳等[12]发现,碳氮比(C/N)能够显著影响Proteobacteria的相对丰度,在C/N为0时,Proteobacteria的相对丰度最低(40.7%),而C/N为5时,Proteobacteria的相对丰度最高(65.2%)。此外,Proteobacteria可被进一步划分为5个纲,分别为α、β、γ、δ和ε。其中,α-、β-、γ-和δ-Proteobacteria普遍共存于营养物去除生化系统中,且β-Proteobacteria是最主导的菌纲[13-14]。然而,本研究中α-、β-、γ-和δ-Proteobacteria的相对丰度分别为58.3%、16.1%、5.9%和1.9%。α-和β-Proteobacteria是较为优势的菌纲,这2类菌纲包括较多的好氧和兼性菌,可以代谢NO2−-N无机化合物。
图2(b)为微生物属水平上物种的组成及相对丰度。硝化杆菌属(Nitrobacter)、中慢生根瘤菌属(Mesorhizobium)、红长命菌属(Rubrivivax)、慢生根菌属(Bradyrhizobium)、伯克氏菌属(Burkholderia)、芽单胞菌属(Gemmatimonas)和丰祐菌属(Opitutus)等相对丰度>1.0%菌群有15属,共占总生物量的60.7%。此外,相对丰度小于<1.0%的菌属主要有链球菌属(Streptococcus)、噬酸菌属(Acidovorax)和鞘氨酸单胞菌属(Sphingomonas)等5属,共占总生物量的4.5%,共同构成了微生物在属水平上的群落结构。在这些微生物菌属中,Nitrobacter、Mesorhizobium和Rubrivivax是占总生物量比例较大的主导优势菌属,平均相对丰度分别为37.8%、3.9%和2.6%。其它微生物菌属的相对丰度总和占了全部生物量的34.9%,也是生化系统中微生物组成的重要部分。
本实验检测出3种NOB菌属,分别为硝化杆菌属(Nitrobacter)、硝化螺菌属(Nitrospira)和硝化刺菌属(Nitrospina)。其中,Nitrobacter是活性污泥系统中最优势菌属(相对丰度为37.8%),而Nitrospira和Nitrospina的相对丰度较低,分别仅为0.03%和0.05%。可见,以NaNO2为唯一氮源和能源,控制运行条件,逐步提高NO2−浓度梯度,可实现NOB种群的富集和优化,并使Nitrobacter成为NOB优势菌属。
有研究表明,Nitrobacter和Nitrospira是生物脱氮系统中最为代表性的NOB菌属[15],但2类菌属的生理生化特性有显著差异。在高NO2−基质底物浓度、高溶解氧条件下,Nitrobacter含量远高于Nitrospira,即有利于Nitrobacter的增殖[16-17]。本实验NOB富集过程中NO2−浓度从10 mg·L−1逐步提高至700 mg·L−1,且运行过程DO浓度维持在4.0 mg·L−1以上,有利于Nitrobacter的培养。相对于Nitrobacter,Nitrospira具有生长速率缓慢、高产率和高底物亲和力等生理生化特性,适宜在基质浓度限制的寡营养环境中生长[18]。姚倩等[19]采用SBR反应器,在NO2−浓度始终低于2 mg·L−1、温度为26 ℃、DO为3~4 mg·L−1的条件下,成功富集了以Nitrospira(相对丰度75%)为主的活性污泥。基于上述分析,本实验的运行效果与SEM及高通量微生物多样性测序结果相一致,充分表明富集培养后SBR系统污泥微生物中NOB菌群中Nitrobacter占绝对优势地位。
-
微生物Spearman关联网络分析是基于OTU在活性污泥样品中的相对丰度分布,利用相关性分析探寻彼此之间呈现正相关或负相关的微生物类群,进而做出关联网络图[20]。通过网络关联分析,找寻群落成员共同出现(Co-occurrence)或彼此排斥(Co-exclusion)的相互作用模式,从而推断不同微生物类群之间可能的相互“协作”或“竞争”关系[21]。使用R语言,计算活性污泥系统丰度位于前50位优势属之间的Spearman等级相关系数,对其中|Spearman’ρ|>0.8且p<0.01的相关优势属构建关联网络,导入Cytoscape进行数据可视化(图3)。图中颜色相同的节点代表这类微生物属于同一个属分类水平,且节点越大,表明该微生物在微生物菌群中的相对丰度越高。节点连线越多,说明系统微生物菌群与该目标微生物之间联系越密切。节点之间的连线表明两者之间存在相关性(红线代表正相关,绿线代表负相关)。
在优势物种关联网络图中,左圆和右圆内微生物菌群均为呈正相关性的菌种,两圆之间均为呈现负相关性的菌种。其中,Nitrobacter中的维氏硝酸杆菌种(Nitrobacter winogradskyi)、汉氏硝酸杆菌种(Nitrobacter hamburgensis)、红假单胞菌种(Rhodopseudomonas)、中慢生根瘤菌种(Mesorhizobium)、生丝微菌种(Hyphomicrobium)及短根瘤菌种(Brad yrhizobium)等表现为正相关关系;但与芽单胞菌种(Gemmatimonas)、纤丝菌种(Leptothrix)、链球菌种(Streptococcus)、分枝杆菌种(Ramlibacter)及寡养单胞菌种(Stenotrophomonas)呈现负相关关系。在所有菌种中,维氏硝酸杆菌种(Nitrobacter winogradskyi)的相对丰度最高,为33.0%,其次为汉氏硝酸杆菌种(Nitrobacter hamburgensis),相对丰度为4.5%,这两个菌种作为Nitrobacter进化树的重要分支,在活性污泥生物脱氮过程扮演重要的角色,它们之间具有较高的相似度(p<0.01)[22]。与Nitrobacter winogradskyi和Nitrobacter hamburgensis呈正相关的主要菌种包括,Mesorhizobium、Bradyrhizobium、Sinorhizobium、Rhodobacter和 Chelativorans等16个菌种。其中,Mesorhizobium与两类菌种的正相关性最强,Spearman’ρ达到0.999和0.998,这表明这些微生物的成员在系统中以协同作用(Co-occurrence)出现,完成NO2−的氧化。与Nitrobacter winogradskyi和Nitrobacter hamburgensis菌种呈负相关的菌种有Variovorax、Ralstonia、Truepera、Streptococcus和Stenotrophomonas等14个菌种。其中,Streptococcus与Variovorax和Ralstonia的负相关性最强,Spearman’ρ达到-0.999和-0.999,表明两类菌种与这些微生物的成员在系统中以彼此排斥(Co-exclusion)的相互作用模式共存,不利于NO2−的氧化。
-
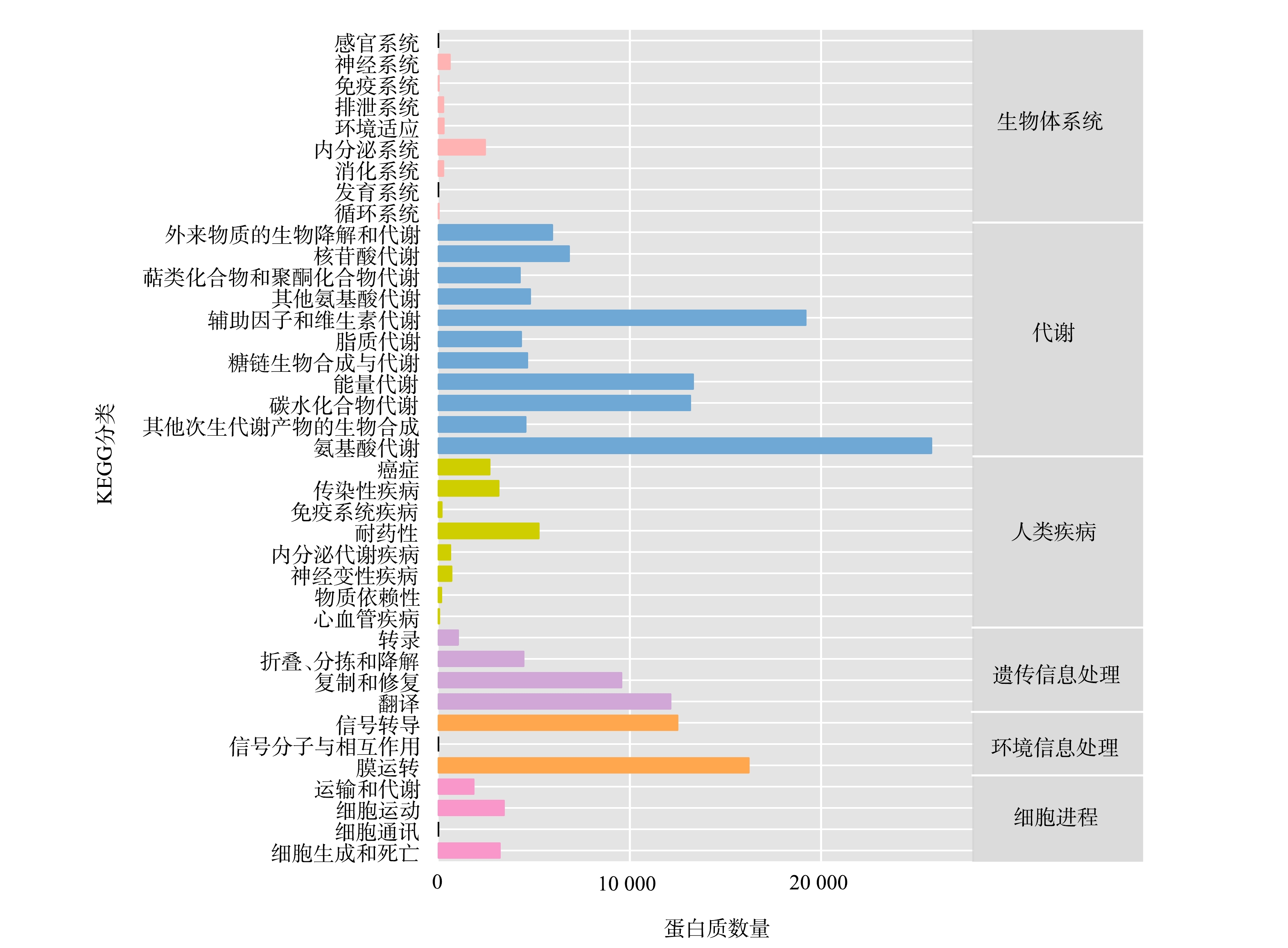
微生物宏基因组基因功能注释是将非冗余蛋白序列集与常用蛋白数据库比对,对各污泥样品中的基因功能进行注释分析。目前,应用最广泛的提供功能注释的数据库是KEGG代谢通路数据库。KEGG为生物代谢通路分析数据库,包括代谢、遗传信息处理、环境信息处理、细胞进程、生物体系统和人类疾病等6大类代谢通路。将蛋白序列和KEGG代谢通路数据库比对,对宏基因组预测得到的基因进行注释和分类。将上述非冗余蛋白序列集上传至KAAS[23]进行功能注释,对注释结果进行汇总,获取各等级的注释结果及对应的功能蛋白绝对丰度信息。
注释结果如图4所示。图中横坐标为蛋白序列注释到相应代谢通路的蛋白质数量,纵坐标左侧为KEGG第二等级代谢通路子功能,其所属的第一等级分类在右侧列出。在第一等级代谢通路中代谢、遗传信息处理、环境信息处理、细胞进程、生物体系统和人类疾病注释到的蛋白质数量分别为106 698、27 149、28 711、8 460、3 804和12 638个。其中,注释到代谢通路的蛋白质数量最多,占总蛋白数量的56.9%,其次为是环境信息处理(15.4%)和遗传信息处理(14.5%),其余3种通路的相应功能的蛋白丰度较低(2.0~6.7%)。新陈代谢功能模块中编码相应功能的基因在功能上的多样性和数量上均具有显著的优势性,反映了微生物新陈代谢是各类菌属与环境之间主导的物质和能量交换以及生物体内物质和能量的生命活动过程。
在第二等级代谢通路子功能水平上,代谢通路中注释到氨基酸代谢的蛋白数值最多,为25 756个(13.7%),其次为辅助因子和维生素代谢,其功能蛋白数量为19 196个(10.2%)。环境信息处理中注释到膜运转的蛋白数量为16 212个(8.6%)。注释到碳水化合物代谢、能量代谢、翻译和信号传导的蛋白质数量几乎处在一个相同的水平,均值为12 771个(6.8%)。基于KEGG的蛋白功能注释功能表明,活性污泥系统内具有相对特定功能的微生物种群。Nitrobacter菌属作为系统中最优势菌种,在将NO2−氧化成NO3−的生化反应过程中,进行着诸多活跃的与新陈代谢相关活动。
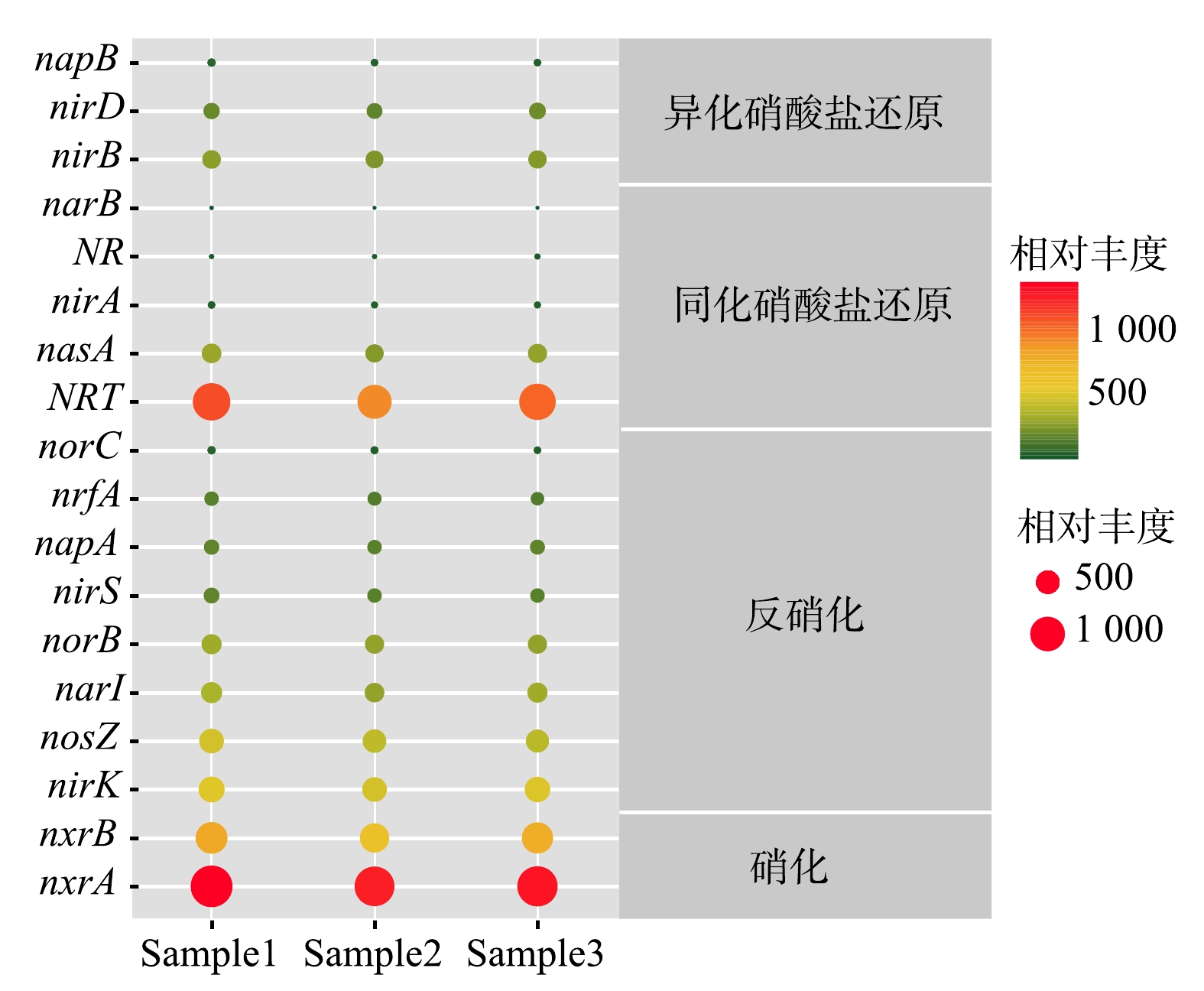
此外,在KEGG第四等级对应的代谢通路上获得各个KO直系同源基因簇的具体注释信息,将所有KO数据映射到KEGG的氮代谢通路图,获得系统与氮代谢有关的酶。在硝化、反硝化、同化硝酸盐还原和异化硝酸盐还原氮转化过程中,共获得18种氮代谢相关的基因(表1、图5)。其中,亚硝酸盐氧化还原酶Nxr(nxrA和nxrB基因)的相对丰度最高,为13.5%。Nxr是NOB的功能标记物,是亚硝酸盐氧化的关键酶,主要有细胞质型和细胞周质型2种类型,都属于络合物铁-硫钼酶家族,由α亚基(nxrA)、β亚基(nxrB)和γ亚基(nxrC)组成[24]。目前,nxrA和nxrB分别作为硝化杆菌(Nitrobacter)和硝化螺菌(Nitrospira)的功能基因和系统发育标记物被广泛应用[25-26],本实验过程所有样品中,均检测出nxrA和nxrB,两者的相对丰度分别为8.6%和4.9%,且在所有的基因中,nxrA的相对丰度最大,这也从功能基因层面证实了Nitrobacter是生化系统内的最优势菌属,亚硝酸盐氧化反应是系统内起主导作用的生化反应。
-
1) 基于16S rRNA宏基因组对活性污泥物种注释和丰度结果显示,Proteobacteria为活性污泥系统中最优势菌门(约占82.2%),硝化杆菌属(Nitrobacter)为最优势菌属(约占37.8%),其余菌属占比均低于5.0%,表明经过富集培养后SBR活性污泥系统微生物群落中NOB占绝对优势地位。
2) 微生物种水平优势物种关联网络表明维氏硝酸杆菌(Nitrobacter winogradskyi)和汉氏硝化细菌(Nitrobacter hamburgensis)相对丰度较高,分别为33.0%、4.5%,是Nitrobacter的主导物种,且两者呈现出显著正相关性。
3) 基于KEGG数据库蛋白功能注释分析结果表明,NOB富集系统进行着多种新陈代谢功能(56.9%)、环境信息处理(15.4%)以及遗传(14.5%)活动,且基于氮代谢过程相关酶的种类及丰度表明nxrA和nxrB是亚硝酸盐氧化的最优势功能基因,两者的相对丰度占全功能基因丰度总和的13.5%。
基于宏基因组学的亚硝酸盐氧化菌属的新陈代谢功能分析
Metabolic function analysis of nitrite oxidizing bacteria based on metagenomics
-
摘要: 为探究亚硝酸盐氧化菌(NOB)的菌群结构及其新陈代谢功能特性,在富集NOB菌属的基础上,基于宏基因组学技术探究了富集活性污泥系统内NOB微生物种群结构、相互作用关系、氮转化相关功能基因及蛋白质功能特性。结果表明,Nitrobacter为富集后活性污泥系统中最优势菌属,相对丰度为37.8%。微生物菌种之间表现出各种错综复杂的关联特征,其中,维氏硝酸杆菌种(Nitrobacter winogradskyi)和汉氏硝化菌种(Nitrobacter hamburgensis)呈现出显著正相关性。此外,新陈代谢是系统内各类菌属中主导的生命活动过程,亚硝酸盐氧化还原酶Nxr为生化系统内氮代谢的关键酶(占比13.5%)。本研究结果可为NOB菌属的富集培养提供参考。Abstract: In order to explore the community structure and metabolic function of nitrite oxidizing bacteria (NOB) in an activated sludge system, the microbial population structure, interaction relationship, nitrogen conversion related functional factors and protein functional characteristics in the NOB enriched activated sludge system were explored based on metagenomics technology. Results showed that Nitrobacter was the most abundant bacteria in the activated sludge system after enrichment, with relative abundance of 37.8%. Among them, Nitrobacter winogradskyi and Nitrobacter hamburgensis showed a strong positive correlation. In addition, metabolism was the most dominant process of all kinds of bacteria in the system, and Nitrite oxidoreductase Nxr was the key enzyme of nitrogen metabolism in biochemical system (accounting for 13.5%). The results of this study can provide a reference for the enrichment culture of NOB bacteria.
-
焦化废水是炼焦工业中煤气净化及焦化产品制作回收等环节产生的混合工业废水[1]。焦化废水水质属性复杂多变,含有大量氨氮、苯酚、氰化物、硫氰化物等特征污染物,并且具有难降解、生物毒性强及生化性差等的特点[2-3]。工业水处理中常用的生物工艺有缺氧/好氧(A/O)工艺、厌氧/缺氧/好氧(A2/O)工艺[4-6]等,但这些工艺在处理焦化废水等高毒性的工业废水时,时常出现硝化失败的情况,其除碳脱氮效果不理想,出水水质不稳定[7-9]。针对上述问题,迫切需要开发更加先进高效的除碳脱氮工艺应用于毒性高的工业废水处理。
好氧-水解-好氧(OHO)作为一种新型脱氮工艺,其原理方面的研究以及工业应用已经证明,该工艺可以很好地解决传统生物工艺除碳脱氮效果不理想、出水水质不稳定的问题[10-12]。OHO工艺具有功能明确的组成单元,与传统工艺相比,采用流化床的三泥法运行模式可以选择性富集微生物群落,并可实现良好的工艺稳定性,突破了高毒性及高COD/TN比废水处理的负荷瓶颈,而且节能降耗。OHO工艺采用基于流化床的三污泥法运行模式,通过回流与超越操作手段,可以实现污泥生物量的富集与微生物功能/丰度调控,活性污泥与焦化废水中的污染物完全混合并充分接触,高效的混合与传质克服了毒性抑制问题。O1单元中好氧微生物氧化去除氰化物、硫氰酸盐、硫化物、苯酚等还原性物质,可大幅度削减废水毒性,为后续工艺单元提供良好的水质条件。毒性降低的焦化废水在H单元进行高效的反硝化脱氮。H单元采用长时间停留,具有容纳毒性负荷的能力,超越比例的控制以H单元中的微生物活性不受进水抑制为基础,与传统工艺相比,降低了厌氧单元的毒性负荷,表现出OHO-MBR工艺的先进性。潘建新[13]发现,异养菌在O1单元富集,自养硝化菌在O2单元富集,O1单元中相对丰度最高的菌属Comamonas贡献有机物的去除,O2单元中AOB菌也具有相当高的相对丰度,贡献了O2单元的硝化,体现了OHO工艺在稳定处理焦化废水时具有的差异化优势。
随着我国污/废水排放标准要求的不断提高,为实现废水零排放的目标,对焦化废水的有效处理、达标排放提出了更加严峻的挑战。膜生物反应器(membrane bioreactor, MBR)是生物处理与膜分离的有机结合,其优质的生物出水水质为后续的脱盐等深度处理营造了优越、稳定的水质条件,成为现有活性污泥处理工艺升级改造、实现废水零排放所优先考虑的生物处理工艺[14-15]。SUN等[16]采用厌氧-缺氧-好氧膜生物反应器(A2O-MBR)处理纺织助剂废水,在内循环比为1.5时,COD、
${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ 基于上述研究结果,本研究以OHO新型生物处理工艺为基础[11],在二级好氧池内加装膜组件,构建了OHO-MBR组合工艺,研究了其处理实际焦化废水的可行性;通过分析工艺对COD、
${\rm{NH}}_4^ + $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_3^ - $ 1. 材料与方法
1.1 实验废水
本研究的OHO-MBR系统进水采用宝武集团广东韶关钢铁有限公司焦化厂混凝预处理后的实际焦化废水。系统进水耗氧有机物(以COD计)质量浓度为2 600~3 477 mg·L−1,进水BOD为1 025~1 440 mg·L−1,进水SCN−-N质量浓度为53~156 mg·L−1,进水TN质量浓度为56~144 mg·L−1,进水
${\rm{NH}}_4^ + $ ${\rm{NO}}_3^ - $ ${\rm{NO}}_2^ - $ 1.2 OHO-MBR系统装置
实验所用连续流OHO-MBR小试装置如图1所示。OHO-MBR反应器槽体为矩形结构,尺寸为814 cm×250 cm×500 cm,总有效容积为56.8 L。该实验系统由O1、H、O2单元组成,采用连续流进水模式。其中O1为一级好氧单元,有效容积20.4 L,分离区5.3 L,通过进水泵调节进水流量,控制进出水平衡,设置曝气装置,保证曝气量在2~3 L·min−1;H为水解单元,有效容积为18.4 L,分离区为5.0 L,通过机械搅拌及水流提升作用,均匀单元水质,维持厌氧环境;O2为二级好氧膜分离单元,有效容积为18.0 L,分离区为4.7 L,调节进气口曝气器保证混合液均匀充氧,膜组件采用沉没式平板膜,有效膜面积为0.10 m2,孔径为0.10 μm,额定膜通量为20 L·(m2·h)−1。各单元主要运行参数见表1。
表 1 OHO-MBR各单元主要运行参数Table 1. Main operating parameters of OHO-MBR.工艺单元 HRT/h SRT/d MLSS/(mg·L−1) pH DO/(mg·L−1) O1 20 20 4 000~6 000 8.0~8.5 4~6 H 18 90 3 000~5 000 7.4~8.1 <0.2 O2-MBR 18 50 4 000~5 000 7.9~8.2 4~6 OHO-MBR反应器运行模式可以为推流、超越与回流及其结合的模式。33%的进水经蠕动泵抽取超越至H单元,回流泵从O2分离区吸取上清液至H单元,回流比R2为0.66。O1/H组合形成高效除碳氨化单元,焦化废水进水经O1好氧单元充分去除有机污染物,有机氮、氰化物和硫氰化物等的含氮化合物均被氧化降解释放出氨氮,随后进入H单元,在水解酸化作用下提升反应器内残余有机污染物的可生化性。H/O2形成一个高效生物脱氮体系,在反硝化作用下,降低废水中的总氮,MBR膜组件对好氧曝气处理后的废水污泥混合液进行高效固液分离,生物单元处理后的废水经膜组件过滤后排出。分别从O1、H、O2单元中定时采集水样,简单沉淀后,取上清液进行检测分析,分析COD、
${\rm{NH}}_4^ + $ 1.3 分析检测
选取SCN−-N、
${\rm{NH}}_4^ + $ ${\rm{NO}}_3^ - $ ${\rm{NO}}_2^ - $ ${\rm{NH}}_4^ + $ ${\rm{NO}}_3^ - $ ${\rm{NO}}_2^ - $ 2. 结果与讨论
2.1 OHO-MBR对SCN−-N的去除效果
图2和图3反映了OHO-MBR反应系统3个处理单元中SCN−-N质量浓度及各单元对污染物去除的贡献情况。在OHO-MBR的进水SCN−-N质量浓度为53.1~156 mg·L−1,除进水波动、DO不足的情况之外,系统出水SCN−-N质量浓度在1.00 mg·L−1以下,表明系统对SCN−-N具有高效的转化效率。
进水SCN−-N在一级好氧单元O1中经微生物作用氨化降解为
${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ 在水解单元H中,缺氧条件下,SCN−-N在水解酶的作用下完全降解。H单元中SCN−-N来源于O1单元出水和超越至H单元的进水之和,由于O2单元回流硝化液会稀释H单元进水中SCN−-N的质量浓度。在58~87 d,由于进水SCN−-N质量浓度大幅增加,导致超越至H单元的SCN−-N质量浓度增加,在回流体积不变的条件下,H单元中SCN−-N质量浓度出现峰值。在系统进水负荷降低后,H单元出水SCN−-N质量浓度由45.7 mg·L−1降至7.49 mg·L−1。
二级好氧单元O2中,
${\rm{NH}}_4^ + $ ${\rm{NO}}_3^ - $ SCN−-N作为还原性物质在废水COD值测定中产生1.10的当量贡献,高浓度SCN−-N为焦化废水的主要耗氧有机物(以COD计)来源,常与酚类和硫化物被称为焦化废水COD三大主要来源之一,故SCN−-N去除效果直接影响出水水质[19-20]。有研究[21]表明,焦化废水中SCN−-N有COS−和CNO− 2种生物降解途径,在不同菌属的作用下降解为相同的最终产物。黄会静[22]发现,20%接种量下SCN−-N降解速率最为合适,且微生物降解难降解污染物质均有一个适应期,适应期的长短与污染物的初始浓度、菌种驯化周期、菌量及培养条件有关。这也解释了在系统启动初期O2单元出水SCN−-N平均质量浓度为2.76 mg·L−1的原因。总体而言,SCN−-N主要在OHO-MBR的好氧单元被转化,在稳定运行情况下,SCN−-N平均去除率为98.5%,基本不构成对生物脱氮的毒性抑制。
2.2 OHO-MBR对
${\rm{NH}}_4^ + $ 图4反映了OHO-MBR反应系统3个处理单元中
${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ 在好氧条件下,焦化废水中的
${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ 水解单元H中,有机氮化合物、氰化物及硫氰化物发生水解氨化作用,有利于后续生物脱氮的进行。稳定运行情况下,H单元
${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ 前序单元进水中残留的
${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ 总体而言,
${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ 2.3 OHO-MBR中对
$ {\bf{NO}}_x^ - $ 图5、图6反映了OHO-MBR反应系统3个处理单元中NOx−-N (
${\rm{NO}}_3^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_3^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_3^ - $ ${\rm{NO}}_3^ - $ ${\rm{NO}}_3^ - $ ${\rm{NH}}_4^ + $ ${\rm{NO}}_3^ - $ 水解单元H是发生反硝化脱氮的主要场所。O1单元出水和O2单元回流液汇入H单元,为H单元提供了
${\rm{NO}}_3^ - $ ${\rm{NO}}_3^ - $ ${\rm{NO}}_3^ - $ ${\rm{NO}}_3^ - $ O2是好氧硝化单元,废水中残余氰化物、硫氰化物、氨氮等污染物在好氧环境中充分降解。在1~3 d内,处于反应器启动初期,污染物在O1中即完成硝化作用,因此,O2单元出水中
${\rm{NO}}_3^ - $ ${\rm{NO}}_3^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_2^ - $ ${\rm{NH}}_4^ + $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_3^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_2^ - $ ${\rm{NO}}_2^ - $ 反硝化是一个涉及到多种酶参与、电子传递及能量转化的过程。本研究中O2到H单元的回流比为66%,可通过提高回流比R2的方式提高OHO-MBR工艺中的TN去除。
2.4 OHO-MBR对TN的去除效果
图7和图8反映了OHO-MBR反应系统3个处理单元中TN质量浓度及各单元对污染物去除的贡献情况。本研究中采用了较低的O2硝化液回流比(66%);但即使在低回流比条件下,TN平均去除率仍可达到65.4%。
TN的去除途径有微生物同化作用和硝化-反硝化作用。好氧单元O1及O2去除TN在于微生物的同化作用;H单元去除TN在于反硝化作用,反硝化的程度决定了OHO-MBR工艺对的TN去除性能。一级好氧单元O1中,SCN−-N降解为
${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ 水解单元H中,回流的好氧硝化液在此单元内经反硝化作用达到脱氮的目标。稳定运行条件下,H单元对去除TN的贡献最高为84.05%。SCN−-N、
${\rm{NH}}_4^ + $ ${\rm{NO}}_3^ - $ ${\rm{NO}}_3^ - $ 在稳定运行状态下,二级好氧单元O2出水中TN平均质量浓度为47.3 mg·L−1,表明OHO-MBR工艺出水中仍含有一定量的TN。O2单元出水中
${\rm{NO}}_3^ - $ ${\rm{NH}}_4^ + $ ${\rm{NO}}_3^ - $ 2.5 OHO-MBR对COD的去除效果
图9和图10反映了OHO-MBR反应系统3个处理单元中COD的变化和各单元对COD的去除贡献。在稳定运行状态下,OHO-MBR的进水COD为2 711~3 477 mg·L−1,O2单元出水COD最低为297 mg·L−1。焦化废水耗氧有机物(以COD计)来源于水中的易生物降解物质和难生物降解物质。COD主要通过好氧条件对还原性物质的氧化及反硝化过程中对有机物的利用来去除。一级好氧单元O1在稳定运行条件下,O1单元出水中COD平均值为491 mg·L−1,O1单元对COD平均去除贡献为84.1%。进水中耗氧有机物(以COD计)主要由有机污染物、酚类、硫氰化物、氰化物、硫化物构成,易降解耗氧有机物(以COD计)在O1单元去除,削减了水中的COD。COD的去除率取决于污泥浓度、溶解氧与进水有机负荷。
H单元通过反硝化过程中对碳源的利用实现对COD的去除。O1单元出水及超越至H单元的进水提供了反硝化所需碳源,O2单元好氧硝化液回流提供
${\rm{NO}}_3^ - $ 二级好氧单元O2在稳定运行条件下,进水COD平均值为1 135 mg·L−1,出水平均值为435 mg·L−1,O2单元对COD去除贡献平均为38.1%。O2单元接纳H单元出水,进一步氧化焦化废水中残留的有机物和还原性物质,从而实现对COD的去除。在47~49 d内,O2单元出水COD上升至1 016 mg·L−1,这是因为O1单元DO不足,导致对有机物的降解能力下降,超出O2单元的COD接纳负荷。MBR作为出水单元可去除悬浮组分及胶体组分,有效截留单元内的活性污泥。WEI等[27]发现,在焦化废水生物处理出水平均残余COD为168.8 mg·L−1的条件下,采用膜过滤(0.45 μm)工艺深度处理后的出水平均COD降为95.1 mg·L−1,膜过滤对生物处理出水COD的平均去除率为43.7%。在稳定运行状态下,经过OHO-MBR工艺处理后,出水COD最低为297 mg·L−1,对COD的最高去除率为89.8%。残余COD主要由可溶性有机物构成[28],可采用混凝沉淀、臭氧氧化或吸附等后物化处理工艺进一步去除,实现出水的排放标准或达到脱盐的入膜水质要求。
2.6 OHO-MBR工艺效能比较
表2所示为不同处理工艺对焦化废水的处理效能对比,其中AO、A2O、A2O-MBR为实验室规模,OHO-MBR为小试装置,OHO为工程应用。OHO设置双好氧单元,可以有效消除或减轻焦化废水的毒性抑制作用,以保证硝化反硝化反应的顺利进行,提高工艺脱氮效率。传统生物工艺如AO和A2O工艺虽然通过前置厌氧单元去除部分有机物,但高浓度的CN−、SCN−以及苯酚会严重抑制有机物的利用,在焦化废水的实际工程中表现为极度不稳定且难以控制,且受冲击后恢复时间较长等问题。结合OHO的MBR反应器可有效截留和富集不易增殖的微生物,解决稳定性问题。SAHARIAH等[33]采用A2O工艺处理焦化废水,实验结果表明,A单元对COD的去除率仅为2%。本工艺在稳定运行状态下,O1单元对COD平均去除贡献率可达84.1%。本研究作为OHO-MBR工艺的可行性初探,即使在偏低的硝化液回流(0.66)及低环境温度(10~20 ℃)条件下,仍可实现对污染物的良好去除,说明工艺处理效能有可进一步拓展提升的空间。
表 2 不同工艺对焦化废水的处理性能Table 2. Treatment performance of coking wastewater by different processes3. 结论
1) OHO-MBR工艺能够稳定降解焦化废水中的有机物及毒性物质,实现脱氮除碳的目标。在总HRT为56 h、进水COD、TN、
${\rm{NH}}_4^ + $ ${\rm{NH}}_4^ + $ 2) O1单元可实现有机物质及毒性污染物的降解;H单元在反硝化脱氮的同时,亦可水解焦化废水中难降解有机物;O2单元实现完全硝化的同时,亦可实现对残余有机物及还原性无机物的最终氧化去除。
3) OHO-MBR作为生物处理与膜分离的有机结合工艺,可有效截留和富集特种功能微生物,提高工艺污泥负荷和出水的分离效率,对焦化废水表现出良好的脱氮除碳效果,同时系统具备抵抗冲击负荷的能力,可为后续的脱盐等深度处理营造优越、稳定的水质条件。OHO-MBR可作为负荷增强、提高水质的备选技术,在不增加新的构筑物条件下,可用于现有工程的升级改造。
-
表 1 氮代谢过程相关酶的种类及丰度
Table 1. Types and abundances of enzymes related to nitrogen metabolism process
反应过程 相对丰度/% 相关酶 反硝化过程 14.3 nirK、nosZ、narI、norB、nirS、napA、nrfA、norC 硝化过程 13.5 nxrA、nxrB 异化硝酸盐还原过程 6.6 nirB、nirD、napB 同化硝酸盐还原过程 9.3 NRT、nasA、nirA、NR、narB -
[1] TAYLOR A E, MYROLD D D, BOTTOMLEY P J. Temperature affects the kinetics of nitrite oxidation and nitrification coupling in four agricultural soils[J]. Soil Biology and Biochemistry, 2019, 136: 107523. doi: 10.1016/j.soilbio.2019.107523 [2] ZHANG L, SHEN Z, FANG W, et al. Composition of bacterial communities in municipal wastewater treatment plant[J]. Science of the Total Environment, 2019, 689: 1181-1191. doi: 10.1016/j.scitotenv.2019.06.432 [3] DAIMS H, S L, WAGNER M. A New Perspective on Microbes Formerly Known as Nitrite-Oxidizing Bacteria[J]. Trends in Microbiology, 2016: 699-712. [4] 国家环境保护局. 水和废水监测分析方法[M]. 4版. 北京: 中国环境科学出版社, 2002. [5] APHA. Standard methods for the examination of water and wastewater[J]. 20th ed. Washington, D. C. :American Public Health Association, 1998: 80-90. [6] HUSON D H, MITRA S, RUSCHEWEYH H J, et al. Integrative analysis of environmental sequences using MEGAN4[J]. Genome Research, 2011, 21(9): 1552-1560. doi: 10.1101/gr.120618.111 [7] HUSOM D H, AUCH A F, QI J, et al. MEGAN analysis of metagenomics data[J]. Genome Research, 2007, 17(3): 377-386. doi: 10.1101/gr.5969107 [8] 于雪. 温度、pH值和游离亚硝酸对亚硝酸盐氧化菌活性动力学及微生物种群结构影响研究[D]. 兰州: 兰州交通大学, 2019. [9] ZHANG T, SHAO M, YE L. 454 pyrosequencing reveals bacterial diversity of activated sludge from 14sewage treatment plants[J]. The ISME Journal, 2011, 6(6): 1137-1147. [10] FANG D X, ZHAO G, XU X Y, et al. Microbial community structures and functions of wastewater treatment systems in plateau and cold regions[J]. Bioresource Technology, 2018, 249: 684. doi: 10.1016/j.biortech.2017.10.063 [11] 杨浩, 张国珍, 杨晓妮, 等. 典型集雨人饮地区窖水微生物群落多样性及差异解析[J]. 环境科学, 2017, 38(11): 4733-4746. [12] 2] 宁高阳, 牛永健, 李维维, 等. C/N对SBR生物脱氮硝化过程微生物结构的动态影响[J]. 中国环境科学, 2020, 40(05): 2053-2061. doi: 10.3969/j.issn.1000-6923.2020.05.023 [13] SUN H W, SHI W Y, CAI CH J, et al. Responses of microbial structures, functions, metabolic pathways and community interactions to different C/N ratios in aerobic nitrification[J]. Bioresource technology, 2020: 311. [14] PENG Y, ZHU G. Biological nitrogen removal with nitrification and denitrification via nitrite pathway[J]. Applied Microbiology and Biotechnology, 2006, 73(1): 15-26. doi: 10.1007/s00253-006-0534-z [15] CAO J S, ZHANG T, WU Y, et al. Correlations of nitrogen removal and core functional genera in full-scale wastewater treatment plants: Influences of different treatment processes and influent characteristics[J]. Bioresource Technology, 2020, 297: 122455. doi: 10.1016/j.biortech.2019.122455 [16] 董怡君, 王淑莹, 汪传新, 等. 亚硝酸盐氧化菌(NOB)的富集培养与其污泥特性分析[J]. 中国环境科学, 2013, 33(11): 1978-1983. [17] 包鹏, 王淑莹, 马斌, 等. 不同溶解氧间歇曝气对亚硝酸盐氧化菌的影响[J]. 中国环境科学, 2016, 36(9): 2696-2702. doi: 10.3969/j.issn.1000-6923.2016.09.024 [18] VIJAYAN A, JAVADRADHAN R, PILLAI D, et al. Nitrospira as versatile nitrifiers: Taxonomy, ecophysiology, genome characteristics, growth, and metabolic diversity[J]. Journal of Basic Microbiology, 2021(2). [19] 姚倩, 彭党聪, 赵俏迪, 等. 活性污泥中硝化螺菌(Nitrospira)的富集及其动力学参数[J]. 环境科学, 2017, 38(12): 5201-5207. [20] FAUST K, RAES J. Microbial interactions: from networks to models[J]. Nature Reviews Microbiology, 2012, 10(8): 538-550. doi: 10.1038/nrmicro2832 [21] JU F, ZHANG T. Bacterial assembly and temporal dynamics in activated sludge of a full-scale municipal wastewater treatment plant[J]. Isme Journal, 2015, 9(3): 683. doi: 10.1038/ismej.2014.162 [22] TESKE A, ALM E, REGAN J M, et al. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria[J]. Journal of Bacteriology, 1994, 176(21): 6623. doi: 10.1128/jb.176.21.6623-6630.1994 [23] MORIYA Y, ITOH M, OKUDA S, et al. KAAS: an automatic genome annotation and pathway reconstruction server[J]. Nucleic Acids Res, 2007, 35: W182-W185. doi: 10.1093/nar/gkm321 [24] PEATER M, MAIXNER F, BERRY D, et al. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira[J]. Environmental Microbiology, 2014, 16(10): 3055-3071. doi: 10.1111/1462-2920.12300 [25] LAI X S, ZHAO Y Q, PAN F X, et al. Enhanced optimal removal of nitrogen and organics from intermittently aerated vertical flow constructed wetlands: Relative COD/N ratios and microbial responses[J]. Chemosphere, 2020: 244. [26] YIN Y N, YANG CH, GU J, et al. Roles of nxrA-like oxidizers and nirS-like reducers in nitrite conversion during swine manure composting[J]. Bioresource Technology, 2019, 297: 122426. -















 下载:
下载: